Characterization of non-starter lactic acid bacteria from Italian ewe cheeses based on phenotypic, genotypic, and cell wall protein analyses
- PMID: 11319075
- PMCID: PMC92830
- DOI: 10.1128/AEM.67.5.2011-2020.2001
Characterization of non-starter lactic acid bacteria from Italian ewe cheeses based on phenotypic, genotypic, and cell wall protein analyses
Abstract
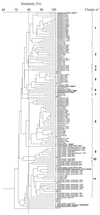
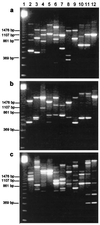
Non-starter lactic acid bacteria (NSLAB) were isolated from 12 Italian ewe cheeses representing six different types of cheese, which in several cases were produced by different manufacturers. A total of 400 presumptive Lactobacillus isolates were obtained, and 123 isolates and 10 type strains were subjected to phenotypic, genetic, and cell wall protein characterization analyses. Phenotypically, the cheese isolates included 32% Lactobacillus plantarum isolates, 15% L. brevis isolates, 12% L. paracasei subsp. paracasei isolates, 9% L. curvatus isolates, 6% L. fermentum isolates, 6% L. casei subsp. casei isolates, 5% L. pentosus isolates, 3% L. casei subsp. pseudoplantarum isolates, and 1% L. rhamnosus isolates. Eleven percent of the isolates were not phenotypically identified. Although a randomly amplified polymorphic DNA (RAPD) analysis based on three primers and clustering by the unweighted pair group method with arithmetic average (UPGMA) was useful for partially differentiating the 10 type strains, it did not provide a species-specific DNA band or a combination of bands which permitted complete separation of all the species considered. In contrast, sodium dodecyl sulfate-polyacrylamide gel electrophoresis cell wall protein profiles clustered by UPGMA were species specific and resolved the NSLAB. The only exceptions were isolates phenotypically identified as L. plantarum and L. pentosus or as L. casei subsp. casei and L. paracasei subsp. paracasei, which were grouped together. Based on protein profiles, Italian ewe cheeses frequently contained four different species and 3 to 16 strains. In general, the cheeses produced from raw ewe milk contained a larger number of more diverse strains than the cheeses produced from pasteurized milk. The same cheese produced in different factories contained different species, as well as strains that belonged to the same species but grouped in different RAPD clusters.
Figures


References
-
- Boot H J, Kolen C P, Pot B, Kersters K, Pouwels P H. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology. 1996;142:2375–2384. - PubMed
-
- British Standard Institution. Chemical analysis of cheese. Part 5. Determination of pH value. British Standard 770. Milton Keyes, United Kingdom: British Standard Institution; 1976.
-
- Collins M D, Phillips B A, Zanoni P. Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int J Syst Bacteriol. 1989;39:105–108.
-
- Corsetti A, Gobbetti M, Smacchi E, De Angelis M, Rossi J. Accelerated ripening of Pecorino Umbro cheese. J Dairy Res. 1998;65:631–642.
-
- Costas M, Pot B, Vandamme P, Kersters K, Owen R J, Hill L R. Interlaboratory comparative study of the numerical analysis of one-dimensional sodium dodecyl sulphate-polyacrylamide gel electrophoretic protein patterns of Campylobacter strains. Electrophoresis. 1990;11:467–474. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

