Kinesin light-chain KLC3 expression in testis is restricted to spermatids
- PMID: 11319135
- PMCID: PMC3161965
- DOI: 10.1095/biolreprod64.5.1320
Kinesin light-chain KLC3 expression in testis is restricted to spermatids
Abstract
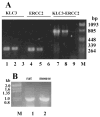
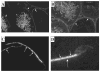
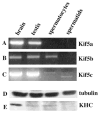
Kinesins are tetrameric motor molecules, consisting of two kinesin heavy chains (KHCs) and two kinesin light chains (KLCs) that are involved in transport of cargo along microtubules. The function of the light chain may be in cargo binding and regulation of kinesin activity. In the mouse, two KLC genes, KLC1 and KLC2, had been identified. KLC1 plays a role in neuronal transport, and KLC2 appears to be more widely expressed. We report the cloning from a testicular cDNA expression library of a mammalian light chain, KLC3. The KLC3 gene is located in close proximity to the ERCC2 gene. KLC3 can be classified as a genuine light chain: it interacts in vitro with the KHC, the interaction is mediated by a conserved heptad repeat sequence, and it associates in vitro with microtubules. In mouse and rat testis, KLC3 protein expression is restricted to round and elongating spermatids, and KLC3 is present in sperm tails. In contrast, KLC1 and KLC2 can only be detected before meiosis in testis. Interestingly, the expression profiles of the three known KHCs and KLC3 differ significantly: Kif5a and Kif5b are not expressed after meiosis, and Kif5c is expressed at an extremely low level in spermatids but is not detectable in sperm tails. Our characterization of the KLC3 gene suggests that it carries out a unique and specialized role in spermatids.
Figures





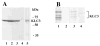
References
-
- Bloom GS, Endow SA. Motor proteins. 1: kinesins. Protein Profile. 1994;1:1059–1116. - PubMed
-
- Beushausen S, Kladakis A, Jaffe H. Kinesin light chains: identification and characterization of a family of proteins from the optic lobe of the squid Loligo pealii. DNA Cell Biol. 1993;12:901–909. - PubMed
-
- Cabeza-Arvelaiz Y, Shih LC, Hardman N, Asselbergs F, Bilbe G, Schmitz A, White B, Siciliano MJ, Lachman LB. Cloning and genetic characterization of the human kinesin light-chain (KLC) gene. DNA Cell Biol. 1993;12:881–892. - PubMed
-
- Fan J, Amos LA. Kinesin light chain isoforms in Caenorhabditis elegans. J Mol Biol. 1994;240:507–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

