Events during initiation of archaeal transcription: open complex formation and DNA-protein interactions
- PMID: 11325929
- PMCID: PMC95201
- DOI: 10.1128/JB.183.10.3025-3031.2001
Events during initiation of archaeal transcription: open complex formation and DNA-protein interactions
Abstract
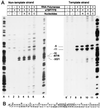
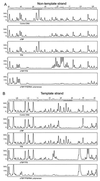
Transcription in Archaea is initiated by association of a TATA box binding protein (TBP) with a TATA box. This interaction is stabilized by the binding of the transcription factor IIB (TFIIB) orthologue TFB. We show here that the RNA polymerase of the archaeon Methanococcus, in contrast to polymerase II, does not require hydrolysis of the beta-gamma bond of ATP for initiation of transcription and open complex formation on linearized DNA. Permanganate probing revealed that the archaeal open complex spanned at least the DNA region from -11 to -1 at a tRNA(Val) promoter. The Methanococcus TBP-TFB promoter complex protected the DNA region from -40 to -14 on the noncoding DNA strand and the DNA segment from -36 to -17 on the coding DNA strand from DNase I digestion. This DNase I footprint was extended only to the downstream end by the addition of the RNA polymerase to position +17 on the noncoding strand and to position +13 on the coding DNA strand.
Figures




References
-
- Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. - PubMed
-
- Carpousis A J, Gralla J D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J Mol Biol. 1985;183:165–177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

