ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity
- PMID: 11325945
- PMCID: PMC95217
- DOI: 10.1128/JB.183.10.3160-3168.2001
ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity
Abstract
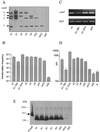
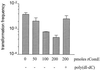
Neisseria gonorrhoeae is naturally able to take up exogenous DNA and undergo genetic transformation. This ability correlates with the presence of functional type IV pili, and uptake of DNA is dependent on the presence of a specific 10-bp sequence. Among the known competence factors in N. gonorrhoeae, none has been shown to interact with the incoming DNA. Here we describe ComE, a DNA-binding protein involved in neisserial competence. The gene comE was identified through similarity searches in the gonococcal genome sequence, using as the query ComEA, the DNA receptor in competent Bacillus subtilis. The gene comE is present in four identical copies in the genomes of both N. gonorrhoeae and Neisseria meningitidis, located downstream of each of the rRNA operons. Single-copy deletion of comE in N. gonorrhoeae did not have a measurable effect on competence, whereas serial deletions led to gradual decrease in transformation frequencies, reaching a 4 x 10(4)-fold reduction when all copies were deleted. Transformation deficiency correlated with impaired ability to take up exogenous DNA; however, the mutants presented normal piliation and twitching motility phenotype. The product of comE has 99 amino acids, with a predicted signal peptide; by immunodetection, a 8-kDa protein corresponding to processed ComE was observed in different strains of N. gonorrhoeae and N. meningitidis. Recombinant His-tagged ComE showed DNA binding activity, without any detectable sequence specificity. Thus, we identified a novel gonococcal DNA-binding competence factor which is necessary for DNA uptake and does not affect pilus biogenesis or function.
Figures




References
-
- Belland R J, Morrison S G, Carlson J H, Hogan D M. Promoter strength influences phase variation of neisserial opagenes. Mol Microbiol. 1997;23:123–135. - PubMed
-
- Chung Y S, Breidt F, Dubnau D. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol Microbiol. 1998;29:905–913. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

