Human testis expresses a specific poly(A)-binding protein
- PMID: 11328870
- PMCID: PMC37253
- DOI: 10.1093/nar/29.9.1872
Human testis expresses a specific poly(A)-binding protein
Abstract
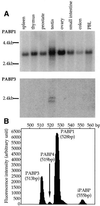
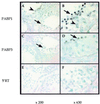
In testis mRNA stability and translation initiation are extensively under the control of poly(A)-binding proteins (PABP). Here we have cloned a new human testis-specific PABP (PABP3) of 631 amino acids (70.1 kDa) with 92.5% identical residues to the ubiquitous PABP1. A northern blot of multiple human tissues hybridised with PABP3- and PABP1-specific oligonucleotide probes revealed two PABP3 mRNAs (2.1 and 2.5 kb) detected only in testis, whereas PABP1 mRNA (3.2 kb) was present in all tested tissues. In human adult testis, PABP3 mRNA expression was restricted to round spermatids, whereas PABP1 was expressed in these cells as well as in pachytene spermatocytes. PABP3-specific antibodies identified a protein of 70 kDa in human testis extracts. This protein binds poly(A) with a slightly lower affinity as compared to PABP1. The human PABP3 gene is intronless with a transcription start site 61 nt upstream from the initiation codon. A sequence of 256 bp upstream from the transcription start site drives the promoter activity of PABP3 and its tissue-specific expression. The expression of PABP3 might be a way to bypass PABP1 translational repression and to produce the amount of PABP needed for active mRNA translation in spermatids.
Figures







References
-
- Yelick P.C., Kwon,Y.H., Flynn,J.F., Borzorgzadeh,A., Kleene,K.C. and Hecht,N.B. (1989) Mouse transition protein 1 is translationally regulated during the postmeiotic stages of spermatogenesis. Mol. Reprod. Dev., 1, 193–200. - PubMed
-
- Steger K., Klonisch,T., Gavenis,K., Drabent,B., Doenecke,D. and Bergmann,M. (1998) Expression of mRNA and protein of nucleoproteins during human spermiogenesis. Mol. Hum. Reprod., 4, 939–945. - PubMed
-
- Schafer M., Nayernia,K., Engel,W. and Schafer,U. (1995) Translational control in spermatogenesis. Dev. Biol., 172, 344–352. - PubMed
-
- Penttila T.L., Yuan,L., Mali,P., Hoog,C. and Parvinen,M. (1995) Haploid gene expression: temporal onset and storage patterns of 13 novel transcripts during rat and mouse spermiogenesis. Biol. Reprod., 53, 499–510. - PubMed
-
- Tafuri S.R., Familari,M. and Wolffe,A.P. (1993) A mouse Y box protein, MSY1, is associated with paternal mRNA in spermatocytes. J. Biol. Chem., 268, 12213–12220. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

