Identification of the 6-sulfate binding site unique to alpha-subunit-containing isozymes of human beta-hexosaminidase
- PMID: 11331008
- PMCID: PMC2910086
- DOI: 10.1021/bi0029200
Identification of the 6-sulfate binding site unique to alpha-subunit-containing isozymes of human beta-hexosaminidase
Abstract
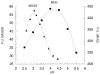
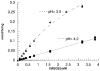
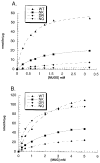
In humans, beta-hexosaminidase A (alphabeta) is required to hydrolyze GM2 ganglioside. A deficiency of either the alpha- or beta-subunit leads to a severe neurological disease, Tay-Sachs or Sandhoff disease, respectively. In mammals beta-hexosaminidase B (betabeta) and S (alphaalpha) are other major and minor isozymes. The primary structures of the alpha- and beta-subunits are 60% identical, but only the alpha-containing isozymes can efficiently hydrolyze beta-linked GlcNAc-6-SO(4) from natural or artificial substrates. Hexosaminidase has been grouped with glycosidases in family 20. A molecular model of the active site of the human hexosaminidase has been generated from the crystal structure of a family 20 bacterial chitobiase. We now use the chitobiase structure to identify residues close to the carbon-6 oxygen of NAG-A, the nonreducing beta-GlcNAc residue of its bound substrate. The chitobiase side chains in the best interactive positions align with alpha-Asn(423)Arg(424) and beta-Asp(453)Leu(454). The change in charge from positive in alpha to negative in beta is consistent with the lower K(m) of hexosaminidase S, and the much higher K(m) and lower pH optimum of hexosaminidase B, toward sulfated versus unsulfated substrates. In vitro mutagenesis, CHO cell expression, and kinetic analyses of an alphaArg(424)Lys hexosaminidase S detected little change in V(max) but a 2-fold increase in K(m) for the sulfated substrate. Its K(m) for the nonsulfated substrate was unaffected. When alphaAsn(423) was converted to Asp, again only the K(m) for the sulfated substrate was changed, increasing by 6-fold. Neutralization of the charge on alphaArg(424) by substituting Gln produced a hexosaminidase S with a K(m) decrease of 3-fold and a V(max) increased by 6-fold for the unsulfated substrate, parameters nearly identical to those of hexosaminidase B at pH 4.2. As well, for the sulfated substrate at pH 4.2 its K(m) was increased 9-fold and its V(max) decreased 1.5-fold, values very similar to those of hexosaminidase B obtained at pH 3.0, where its betaAsp(453) becomes protonated.
Figures


References
-
- O’Dowd BF, Klavins MH, Willard HF, Gravel R, Lowden JA, Mahuran DJ. J Biol Chem. 1986;261:12680–12685. - PubMed
-
- Mahuran DJ, Lowden JA. Can J Biochem. 1980;58:287–294. - PubMed
-
- Hou Y, Tse R, Mahuran DJ. Biochemistry. 1996;35:3963–3969. - PubMed
-
- Gravel RA, Clarke JTR, Kaback MM, Mahuran D, Sandhoff K, Suzuki K. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. McGraw-Hill; New York: 1995. pp. 2839–2879.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

