Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions
- PMID: 11331577
- PMCID: PMC125439
- DOI: 10.1093/emboj/20.9.2111
Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions
Abstract
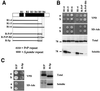
The nuclear-encoded Sup35p protein is responsible for the prion-like [PSI(+)] determinant of yeast, with Sup35p existing largely as a high molecular weight aggregate in [PSI(+)] strains. Here we show that the five oligopeptide repeats present at the N-terminus of Sup35p are responsible for stabilizing aggregation of Sup35p in vivo. Sequential deletion of the oligopeptide repeats prevented the maintenance of [PSI(+)] by the truncated Sup35p, although deletants containing only two repeats could be incorporated into pre-existing aggregates of wild-type Sup35p. The mammalian prion protein PrP also contains similar oligopeptide repeats and we show here that a human PrP repeat (PHGGGWGQ) is able functionally to replace a Sup35p oligopeptide repeat to allow stable [PSI(+)] propagation in vivo. Our data suggest a model in which the oligopeptide repeats in Sup35p stabilize intermolecular interactions between Sup35p proteins that initiate establishment of the aggregated state. Modulating repeat number therefore alters the rate of yeast prion conversion in vivo. Furthermore, there appears to be evolutionary conservation of function of the N-terminally located oligopeptide repeats in prion propagation.
Figures





Similar articles
-
Oligopeptide-repeat expansions modulate 'protein-only' inheritance in yeast.Nature. 1999 Aug 5;400(6744):573-6. doi: 10.1038/23048. Nature. 1999. PMID: 10448860
-
[Structure and functional similarity of yeast Sup35p and Ure2p proteins to mammalian prions].Mol Biol (Mosk). 1995 Jul-Aug;29(4):750-5. Mol Biol (Mosk). 1995. PMID: 7476941 Review. Russian.
-
[Fusion of glutathione S-transferase with the N-terminus of yeast Sup35p protein inhibits its prion-like properties].Genetika. 1997 May;33(5):610-5. Genetika. 1997. PMID: 9273317 Russian.
-
Is there a human [psi]?C R Acad Sci III. 1996 Jun;319(6):487-92. C R Acad Sci III. 1996. PMID: 8881282
-
[New aspects of research upon the yeast Saccharomyces cerevisiae [PSI+] prion].Postepy Biochem. 2007;53(2):182-7. Postepy Biochem. 2007. PMID: 17969880 Review. Polish.
Cited by
-
Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos.PLoS Genet. 2006 Aug 25;2(8):e134. doi: 10.1371/journal.pgen.0020134. Epub 2006 Jul 13. PLoS Genet. 2006. PMID: 16934003 Free PMC article.
-
Distinct amino acid compositional requirements for formation and maintenance of the [PSI⁺] prion in yeast.Mol Cell Biol. 2015 Mar;35(5):899-911. doi: 10.1128/MCB.01020-14. Epub 2014 Dec 29. Mol Cell Biol. 2015. PMID: 25547291 Free PMC article.
-
Defining the role of the polyasparagine repeat domain of the S. cerevisiae transcription factor Azf1p.PLoS One. 2021 May 21;16(5):e0247285. doi: 10.1371/journal.pone.0247285. eCollection 2021. PLoS One. 2021. PMID: 34019539 Free PMC article.
-
Alternative assembly pathways of the amyloidogenic yeast prion determinant Sup35-NM.EMBO Rep. 2007 Dec;8(12):1196-201. doi: 10.1038/sj.embor.7401096. Epub 2007 Nov 2. EMBO Rep. 2007. PMID: 17975557 Free PMC article.
-
The genetic control of the formation and propagation of the [PSI+] prion of yeast.Prion. 2007 Apr-Jun;1(2):101-9. doi: 10.4161/pri.1.2.4665. Epub 2007 Apr 28. Prion. 2007. PMID: 19164924 Free PMC article. Review.
References
-
- Basler K., Oesch,B., Scott,M., Westaway,D., Walchli,M., Groth,D.F., McKinley,M.P., Prusiner,S.B. and Weissmann,C. (1986) Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell, 46, 417–428. - PubMed
-
- Chernoff Y.O., Derkach,I.L. and Inge-Vechtomov,S.G. (1993) Multicopy SUP35 gene induces de-novo appearance of ψ-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. - PubMed
-
- Chiesa R., Piccardo,P., Ghetti,B. and Harris,D.A. (1998) Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron, 21, 1339–1351. - PubMed
-
- Cox B.S. (1965) Ψ, a cytoplasmic suppressor of super-suppression in yeast. Heredity, 20, 505–521.
-
- DePace A.H., Santoso,A., Hillner,P. and Weissman,J.S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell, 93, 1241–1252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

