Changes in global gene expression patterns during development and maturation of the rat kidney
- PMID: 11331749
- PMCID: PMC33267
- DOI: 10.1073/pnas.091110798
Changes in global gene expression patterns during development and maturation of the rat kidney
Abstract
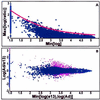
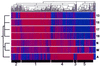
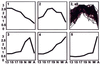
We set out to define patterns of gene expression during kidney organogenesis by using high-density DNA array technology. Expression analysis of 8,740 rat genes revealed five discrete patterns or groups of gene expression during nephrogenesis. Group 1 consisted of genes with very high expression in the early embryonic kidney, many with roles in protein translation and DNA replication. Group 2 consisted of genes that peaked in midembryogenesis and contained many transcripts specifying proteins of the extracellular matrix. Many additional transcripts allied with groups 1 and 2 had known or proposed roles in kidney development and included LIM1, POD1, GFRA1, WT1, BCL2, Homeobox protein A11, timeless, pleiotrophin, HGF, HNF3, BMP4, TGF-alpha, TGF-beta2, IGF-II, met, FGF7, BMP4, and ganglioside-GD3. Group 3 consisted of transcripts that peaked in the neonatal period and contained a number of retrotransposon RNAs. Group 4 contained genes that steadily increased in relative expression levels throughout development, including many genes involved in energy metabolism and transport. Group 5 consisted of genes with relatively low levels of expression throughout embryogenesis but with markedly higher levels in the adult kidney; this group included a heterogeneous mix of transporters, detoxification enzymes, and oxidative stress genes. The data suggest that the embryonic kidney is committed to cellular proliferation and morphogenesis early on, followed sequentially by extracellular matrix deposition and acquisition of markers of terminal differentiation. The neonatal burst of retrotransposon mRNA was unexpected and may play a role in a stress response associated with birth. Custom analytical tools were developed including "The Equalizer" and "eBlot," which contain improved methods for data normalization, significance testing, and data mining.
Figures




Similar articles
-
Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development.Kidney Int. 2003 Dec;64(6):1997-2008. doi: 10.1046/j.1523-1755.2003.00383.x. Kidney Int. 2003. PMID: 14633122
-
D-Serine exposure resulted in gene expression changes indicative of activation of fibrogenic pathways and down-regulation of energy metabolism and oxidative stress response.Toxicology. 2008 Jan 14;243(1-2):177-92. doi: 10.1016/j.tox.2007.10.009. Epub 2007 Oct 23. Toxicology. 2008. PMID: 18061331
-
Temporal and spatial transcriptional programs in murine kidney development.Physiol Genomics. 2005 Oct 17;23(2):159-71. doi: 10.1152/physiolgenomics.00043.2005. Epub 2005 Jul 5. Physiol Genomics. 2005. PMID: 15998744
-
Profiling gene expression in kidney development.Nephron Exp Nephrol. 2004;98(4):e109-13. doi: 10.1159/000081554. Nephron Exp Nephrol. 2004. PMID: 15627793 Review.
-
Profiling gene expression in kidney development.Nephron Exp Nephrol. 2003;95(1):e1-6. doi: 10.1159/000073017. Nephron Exp Nephrol. 2003. PMID: 14520008 Review.
Cited by
-
Postnatal expression of transport proteins involved in acid-base transport in mouse kidney.Pflugers Arch. 2004 Apr;448(1):16-28. doi: 10.1007/s00424-003-1227-4. Epub 2004 Jan 31. Pflugers Arch. 2004. PMID: 14758480
-
The effect of hyaluronic acid size and concentration on branching morphogenesis and tubule differentiation in developing kidney culture systems: potential applications to engineering of renal tissues.Biomaterials. 2007 Nov;28(32):4806-17. doi: 10.1016/j.biomaterials.2007.07.034. Epub 2007 Aug 15. Biomaterials. 2007. PMID: 17706761 Free PMC article.
-
Analysis of metagene portraits reveals distinct transitions during kidney organogenesis.Sci Signal. 2008 Dec 9;1(49):ra16. doi: 10.1126/scisignal.1163630. Sci Signal. 2008. PMID: 19066399 Free PMC article.
-
Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells.Nucleic Acids Res. 2008 Feb;36(2):648-65. doi: 10.1093/nar/gkm1045. Epub 2007 Dec 10. Nucleic Acids Res. 2008. PMID: 18073200 Free PMC article.
-
Organogenesis forum lecture: In vitro kidney development, tissue engineering and systems biology.Organogenesis. 2008 Jul;4(3):137-43. doi: 10.4161/org.4.3.6498. Organogenesis. 2008. PMID: 19279725 Free PMC article.
References
-
- Saxen L. Organogenesis of the Kidney. Cambridge, U.K.: Cambridge Univ. Press; 1987.
-
- Pohl M, Stuart R O, Sakurai H, Nigam S K. Annu Rev Physiol. 2000;62:595–620. - PubMed
-
- Pavlova A, Stuart R O, Pohl M, Nigam S K. Am J Physiol. 1999;277:F650–F663. - PubMed
-
- Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

