Trans-spliced leader addition to mRNAs in a cnidarian
- PMID: 11331766
- PMCID: PMC33275
- DOI: 10.1073/pnas.101049998
Trans-spliced leader addition to mRNAs in a cnidarian
Abstract
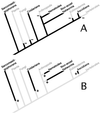
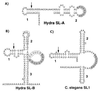
A search of databases with the sequence from the 5' untranslated region of a Hydra cDNA clone encoding a receptor protein-tyrosine kinase revealed that a number of Hydra cDNAs contain one of two different sequences at their 5' ends. This finding suggested the possibility that mRNAs in Hydra receive leader sequences by trans-splicing. This hypothesis was confirmed by the finding that the leader sequences are transcribed as parts of small RNAs encoded by genes located in the 5S rRNA clusters of Hydra. The two spliced leader (SL) RNAs (SL-A and -B) contain splice donor dinucleotides at the predicted positions, and genes that receive SLs contain splice acceptor dinucleotides at the predicted positions. Both of the SL RNAs are bound by antibody against trimethylguanosine, suggesting that they contain a trimethylguanosine cap. The predicted secondary structures of the Hydra SL RNAs show significant differences from the structures predicted for the SLs of other organisms. Messenger RNAs have been identified that can receive either SL-A or -B, although the impact of the two different SLs on the function of the mRNA is unknown. The presence and features of SL addition in the phylum Cnidaria raise interesting questions regarding the evolution of this process.
Figures

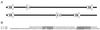

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

