Electronic detection of nucleic acids: a versatile platform for molecular diagnostics
- PMID: 11333303
- PMCID: PMC1907350
- DOI: 10.1016/S1525-1578(10)60655-1
Electronic detection of nucleic acids: a versatile platform for molecular diagnostics
Abstract
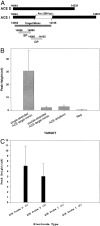
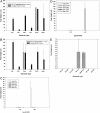
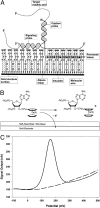
A novel platform for the electronic detection of nucleic acids on microarrays is introduced and shown to perform well as a selective detection system for applications in molecular diagnostics. A gold electrode in a printed circuit board is coated with a self-assembled monolayer (SAM) containing DNA capture probes. Unlabeled nucleic acid targets are immobilized on the surface of the SAM through sequence-specific hybridization with the DNA capture probe. A separate signaling probe, containing ferrocene-modified nucleotides and complementary to the target in the region adjoining the capture probe binding site, is held in close proximity to the SAM in a sandwich complex. The SAM allows electron transfer between the immobilized ferrocenes and the gold, while insulating the electrode from soluble redox species, including unbound signaling probes. Here, we demonstrate sequence-specific detection of amplicons after simple dilution of the reaction product into hybridization buffer. In addition, single nucleotide polymorphism discrimination is shown. A genotyping chip for the C282Y single nucleotide polymorphism associated with hereditary hemochromatosis is used to confirm the genotype of six patients' DNA. In addition, a gene expression-monitoring chip is described that surveys five genes that are differentially regulated in the cellular apoptosis response. Finally, custom modification of individual electrodes through sequence-specific hybridization demonstrates the potential of this system for infectious disease diagnostics. The versatility of the electronic detection platform makes it suitable for multiple applications in diagnostics and pharmacogenetics.
Figures

References
-
- Abramowitz S: DNA Analysis in microfabricated formats. J Biomed Dev 1999, 1:107-112 - PubMed
-
- Zhang MQ: Large-scale gene expression data analysis: a new challenge to computational biologists. Genome Res 1999, 9:681-688 - PubMed
-
- Farkas DH: Bioelectronic detection of DNA and the automation of mol diagnostics. J Assoc Lab Automat 1999, 4:20-24
-
- Creager S, Yu CJ, Bamdad C, O’Connor S, MacLean T, Lam E, Chong Y, Olsen GT, Luo J, Gozin M, Kayyem JF: Electron transfer at electrodes through conjugated “molecular wire” bridges. J Am Chem Soc, 1999, 121:1059–1064
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

