Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches
- PMID: 11333878
- PMCID: PMC114902
- DOI: 10.1128/JVI.75.11.4990-4998.2001
Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches
Abstract
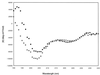
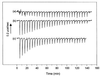
The catalytic subunit, Pol, of herpes simplex virus DNA polymerase interacts via its extreme C terminus with the processivity subunit, UL42. This interaction is critical for viral replication and thus a potential target for antiviral drug action. To investigate the Pol-binding region on UL42, we engineered UL42 mutations but also used random peptide display to identify artificial ligands of the Pol C terminus. The latter approach selected ligands with homology to residues 171 to 176 of UL42. Substitution of glutamine 171 with alanine greatly impaired binding to Pol and stimulation of long-chain DNA synthesis by Pol, identifying this residue as crucial for subunit interactions. To study these interactions quantitatively, we used isothermal titration calorimetry and wild-type and mutant forms of Pol-derived peptides and UL42. Each of three peptides corresponding to either the last 36, 27, or 18 residues of Pol bound specifically to UL42 in a 1:1 complex with a dissociation constant of 1 to 2 microM. Thus, the last 18 residues suffice for most of the binding energy, which was due mainly to a change in enthalpy. Substitutions at positions corresponding to Pol residue 1228 or 1229 or at UL42 residue 171 abolished or greatly reduced binding. These residues participate in hydrogen bonds observed in the crystal structure of the C terminus of Pol bound to UL42. Thus, interruption of these few bonds is sufficient to disrupt the interaction, suggesting that small molecules targeting the relevant side chains could interfere with Pol-UL42 binding.
Figures






References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987.
-
- Bridges K G, Hua Q, Brigham-Burke M R, Martin J D, Hensley P, Dahl C E, Weiss M A, Coen D M. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J Biol Chem. 2000;275:472–478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

