Avian reovirus major mu-class outer capsid protein influences efficiency of productive macrophage infection in a virus strain-specific manner
- PMID: 11333882
- PMCID: PMC114906
- DOI: 10.1128/JVI.75.11.5027-5035.2001
Avian reovirus major mu-class outer capsid protein influences efficiency of productive macrophage infection in a virus strain-specific manner
Abstract
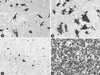
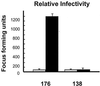
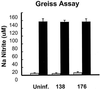
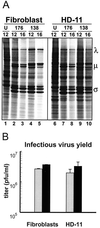
We determined that the highly pathogenic avian reovirus strain 176 (ARV-176) possesses an enhanced ability to establish productive infections in HD-11 avian macrophages compared to avian fibroblasts. Conversely, the weakly pathogenic strain ARV-138 shows no such macrophagotropic tendency. The macrophage infection capability of the two viruses did not reflect differences in the ability to either induce or inhibit nitric oxide production. Moderate increases in the ARV-138 multiplicity of infection resulted in a concomitant increase in macrophage infection, and under such conditions the kinetics and extent of the ARV-138 replication cycle were equivalent to those of the highly infectious ARV-176 strain. These results indicated that both viruses are apparently equally capable of replicating in an infected macrophage, but they differ in the ability to establish productive infections in these cells. Using a genetic reassortant approach, we determined that the macrophagotropic property of ARV-176 reflects a post-receptor-binding step in the virus replication cycle and that the ARV-176 M2 genome segment is required for efficient infection of HD-11 cells. The M2 genome segment encodes the major mu-class outer capsid protein (muB) of the virus, which is involved in virus entry and transcriptase activation, suggesting that a host-specific influence on ARV entry and/or uncoating may affect the likelihood of the virus establishing a productive infection in a macrophage cell.
Figures



Similar articles
-
Characterization of two avian reoviruses that exhibit strain-specific quantitative differences in their syncytium-inducing and pathogenic capabilities.Virology. 1998 Oct 25;250(2):263-72. doi: 10.1006/viro.1998.9371. Virology. 1998. PMID: 9792837
-
Adaptation of reovirus to growth in the presence of protease inhibitor E64 segregates with a mutation in the carboxy terminus of viral outer-capsid protein sigma3.J Virol. 2001 Apr;75(7):3197-206. doi: 10.1128/JVI.75.7.3197-3206.2001. J Virol. 2001. PMID: 11238846 Free PMC article.
-
The sequence and phylogenetic analysis of avian reovirus genome segments M1, M2, and M3 encoding the minor core protein muA, the major outer capsid protein muB, and the nonstructural protein muNS.J Virol Methods. 2006 May;133(2):146-57. doi: 10.1016/j.jviromet.2005.10.031. Epub 2005 Dec 6. J Virol Methods. 2006. PMID: 16337282
-
Reovirus capsid proteins sigma 3 and mu 1: interactions that influence viral entry, assembly, and translational control.Curr Top Microbiol Immunol. 1998;233(Pt 1):167-83. doi: 10.1007/978-3-642-72092-5_8. Curr Top Microbiol Immunol. 1998. PMID: 9599926 Review. No abstract available.
-
Coupling of rotavirus genome replication and capsid assembly.Adv Virus Res. 2007;69:167-201. doi: 10.1016/S0065-3527(06)69004-0. Adv Virus Res. 2007. PMID: 17222694 Review.
Cited by
-
Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction.Virology. 2005 Dec 5;343(1):25-35. doi: 10.1016/j.virol.2005.08.002. Epub 2005 Sep 8. Virology. 2005. PMID: 16153672 Free PMC article.
-
Erythroid Progenitor Cells in Atlantic Salmon (Salmo salar) May Be Persistently and Productively Infected with Piscine Orthoreovirus (PRV).Viruses. 2019 Sep 5;11(9):824. doi: 10.3390/v11090824. Viruses. 2019. PMID: 31491892 Free PMC article.
-
Dynamics of Polarized Macrophages and Activated CD8+ Cells in Heart Tissue of Atlantic Salmon Infected With Piscine Orthoreovirus-1.Front Immunol. 2021 Sep 16;12:729017. doi: 10.3389/fimmu.2021.729017. eCollection 2021. Front Immunol. 2021. PMID: 34603301 Free PMC article.
-
Immunohistochemical detection of piscine reovirus (PRV) in hearts of Atlantic salmon coincide with the course of heart and skeletal muscle inflammation (HSMI).Vet Res. 2012 Apr 9;43(1):27. doi: 10.1186/1297-9716-43-27. Vet Res. 2012. PMID: 22486941 Free PMC article.
-
Proteomics analysis of the DF-1 chicken fibroblasts infected with avian reovirus strain S1133.PLoS One. 2014 Mar 25;9(3):e92154. doi: 10.1371/journal.pone.0092154. eCollection 2014. PLoS One. 2014. PMID: 24667214 Free PMC article.
References
-
- Beug H, von Kirchbach A, Doderlein G, Conscience J F, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. - PubMed
-
- Borsa J, Sargent M D, Kay C M, Oikawa K. Circular dichroism of intermediate particles of reovirus: elucidation of the mechanism underlying the specific monovalent cation effects on uncoating. Biochim Biophys Acta. 1976;451:619–627. - PubMed
-
- Borsa J, Sargent M D, Lievaart P A, Copps T P. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. - PubMed
-
- Bruun T, Kristoffersen A K, Rollag H, Gegre M. Interaction of herpes simplex virus with mononuclear phagocytes is dependent on the differentiation stage of the cells. APMIS. 1998;106:305–314. - PubMed
-
- Clark F D, Ni Y, Collisson E W, Kemp M C. Characterization of avian reovirus strain-specific polymorphisms. Avian Dis. 1990;34:304–314. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

