Primate and feline lentivirus vector RNA packaging and propagation by heterologous lentivirus virions
- PMID: 11333894
- PMCID: PMC114918
- DOI: 10.1128/JVI.75.11.5129-5140.2001
Primate and feline lentivirus vector RNA packaging and propagation by heterologous lentivirus virions
Abstract
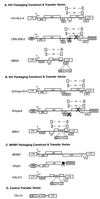
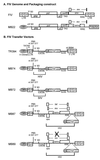
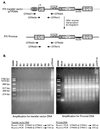
Development of safe and effective gene transfer systems is critical to the success of gene therapy protocols for human diseases. Currently, several primate lentivirus-based gene transfer systems, such as those based on human and simian immunodeficiency viruses (HIV/SIV), are being tested; however, their use in humans raises safety concerns, such as the generation of replication-competent viruses through recombination with related endogenous retroviruses or retrovirus-like elements. Due to the greater phylogenetic distance from primate lentiviruses, feline immunodeficiency virus (FIV) is becoming the lentivirus of choice for human gene transfer systems. However, the safety of FIV-based vector systems has not been tested experimentally. Since lentiviruses such as HIV-1 and SIV have been shown to cross-package their RNA genomes, we tested the ability of FIV RNA to get cross-packaged into primate lentivirus particles such as HIV-1 and SIV, as well as a nonlentiviral retrovirus such as Mason-Pfizer monkey virus (MPMV), and vice versa. Our results reveal that FIV RNA can be cross-packaged by primate lentivirus particles such as HIV-1 and SIV and vice versa; however, a nonlentivirus particle such as MPMV is unable to package FIV RNA. Interestingly, FIV particles can package MPMV RNA but cannot propagate the vector RNA further for other steps of the retrovirus life cycle. These findings reveal that diverse retroviruses are functionally more similar than originally thought and suggest that upon coinfection of the same host, cross- or copackaging may allow distinct retroviruses to generate chimeric variants with unknown pathogenic potential.
Figures


References
-
- Bach F H, Robson S C, Winkler H, Ferran C, Stuhlmeier K M, Wrighton C M, Hancock W W. Barriers to xenotransplantation. Nat Med. 1995;1:869–873. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

