Identification of the regions of Fv1 necessary for murine leukemia virus restriction
- PMID: 11333899
- PMCID: PMC114923
- DOI: 10.1128/JVI.75.11.5182-5188.2001
Identification of the regions of Fv1 necessary for murine leukemia virus restriction
Abstract
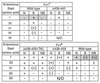
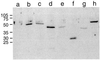
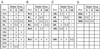
The Fv1 gene restricts murine leukemia virus replication via an interaction with the viral capsid protein. To study this interaction, a number of mutations, including a series of N-terminal and C-terminal deletions, internal deletions, and a number of single-amino-acid substitutions, were introduced into the n and b alleles of the Fv1 gene and the effects of these changes on virus restriction were measured. A significant fraction of the Fv1 protein was not required for restriction; however, retention of an intact major homology region as well as of domains toward the N and C termini was essential. Binding specificity appeared to be a combinatorial property of a number of residues within the C-terminal portion of Fv1.
Figures
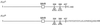




References
-
- Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
-
- Campos-Olivas R, Newman J L, Summers M F. Solution structure and dynamics of the Rous Sarcoma Virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

