Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein
- PMID: 11333914
- PMCID: PMC114938
- DOI: 10.1128/JVI.75.11.5335-5342.2001
Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein
Abstract
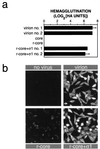
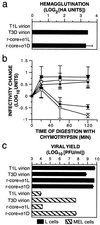
Mammalian reoviruses, prototype members of the Reoviridae family of nonenveloped double-stranded RNA viruses, use at least three proteins--sigma1, mu1, and sigma3--to enter host cells. sigma1, a major determinant of cell tropism, mediates viral attachment to cellular receptors. Studies of sigma1 functions in reovirus entry have been restricted by the lack of methodologies to produce infectious virions containing engineered mutations in viral proteins. To mitigate this problem, we produced virion-like particles by "recoating" genome-containing core particles that lacked sigma1, mu1, and sigma3 with recombinant forms of these proteins in vitro. Image reconstructions from cryoelectron micrographs of the recoated particles revealed that they closely resembled native virions in three-dimensional structure, including features attributable to sigma1. The recoated particles bound to and infected cultured cells in a sigma1-dependent manner and were approximately 1 million times as infectious as cores and 0.5 times as infectious as native virions. Experiments with recoated particles containing recombinant sigma1 from either of two different reovirus strains confirmed that differences in cell attachment and infectivity previously observed between those strains are determined by the sigma1 protein. Additional experiments showed that recoated particles containing sigma1 proteins with engineered mutations can be used to analyze the effects of such mutations on the roles of particle-bound sigma1 in infection. The results demonstrate a powerful new system for molecular genetic dissections of sigma1 with respect to its structure, assembly into particles, and roles in entry.
Figures



Similar articles
-
In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins mu1 and sigma3.J Virol. 1999 May;73(5):3941-50. doi: 10.1128/JVI.73.5.3941-3950.1999. J Virol. 1999. PMID: 10196289 Free PMC article.
-
Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed sigma3 protein: an approach for analyzing sigma3 functions during virus entry.J Virol. 1999 Apr;73(4):2963-73. doi: 10.1128/JVI.73.4.2963-2973.1999. J Virol. 1999. PMID: 10074146 Free PMC article.
-
A monoclonal antibody specific for reovirus outer-capsid protein sigma3 inhibits sigma1-mediated hemagglutination by steric hindrance.J Virol. 2001 Jul;75(14):6625-34. doi: 10.1128/JVI.75.14.6625-6634.2001. J Virol. 2001. PMID: 11413330 Free PMC article.
-
Attachment and cell entry of mammalian orthoreovirus.Curr Top Microbiol Immunol. 2006;309:1-38. doi: 10.1007/3-540-30773-7_1. Curr Top Microbiol Immunol. 2006. PMID: 16909895 Review.
-
From touchdown to transcription: the reovirus cell entry pathway.Curr Top Microbiol Immunol. 2010;343:91-119. doi: 10.1007/82_2010_32. Curr Top Microbiol Immunol. 2010. PMID: 20397070 Free PMC article. Review.
Cited by
-
Reovirus mu1 structural rearrangements that mediate membrane penetration.J Virol. 2006 Dec;80(24):12367-76. doi: 10.1128/JVI.01343-06. Epub 2006 Sep 27. J Virol. 2006. PMID: 17005655 Free PMC article.
-
The delta region of outer-capsid protein micro 1 undergoes conformational change and release from reovirus particles during cell entry.J Virol. 2003 Dec;77(24):13361-75. doi: 10.1128/jvi.77.24.13361-13375.2003. J Virol. 2003. PMID: 14645591 Free PMC article.
-
Reduction of virion-associated σ1 fibers on oncolytic reovirus variants promotes adaptation toward tumorigenic cells.J Virol. 2015 Apr;89(8):4319-34. doi: 10.1128/JVI.03651-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653434 Free PMC article.
-
Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics.J Mol Biol. 2010 Apr 2;397(3):852-63. doi: 10.1016/j.jmb.2009.12.027. Epub 2009 Dec 28. J Mol Biol. 2010. PMID: 20036256 Free PMC article.
-
Disulfide bonding among micro 1 trimers in mammalian reovirus outer capsid: a late and reversible step in virion morphogenesis.J Virol. 2003 May;77(9):5389-400. doi: 10.1128/jvi.77.9.5389-5400.2003. J Virol. 2003. PMID: 12692241 Free PMC article.
References
-
- Armstrong G D, Paul R W, Lee P W K. Studies on reovirus receptors of L cells: virus binding characteristics and comparison with reovirus receptors of erythrocytes. Virology. 1984;138:37–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

