Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis
- PMID: 11337365
- PMCID: PMC1891939
- DOI: 10.1016/S0002-9440(10)64123-5
Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis
Abstract
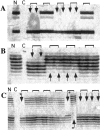
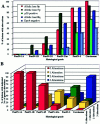
The presumed precursor lesions of pancreatic ductal adenocarcinoma were recently classified according to their increasing grade of dysplasia and were designated as pancreatic intraepithelial neoplasia (PanIN) 1 through 3. In this study, we tested whether molecular genetic alterations can be correlated with this classification and may help to further categorize the various PanIN grades. We determined the frequencies of allelic loss at chromosomal arms 9p, 17p, and 18q in 81 microdissected duct lesions of various PanIN grades, using a combination of whole genome amplification and microsatellite analysis. In addition we examined the p53 and Dpc4 protein expression patterns by immunohistochemical analysis. In PanIN-1, we did not detect allelic losses. In PanIN-2, allelic losses were found in increasing frequency, and were particularly high in those lesions with moderate-grade dysplasia (low grade, 20, 33, and 17%, loss at 9p, 17p, and 18q, respectively; moderate grade, 46, 77, and 58%). PanIN-3 and invasive carcinomas exhibited abundant losses. Abnormal p53 and Dpc4 protein expression was only rarely identified in PanIN-2 lesions, but occurred frequently in PanIN-3 lesions and invasive carcinomas. The combined genetic and protein expression data support a model in which allelic loss is the first hit in the biallelic inactivation of the p53 and DPC4 tumor suppressor genes. In addition, our data indicate that allelic loss analysis may be useful in separating PanIN-2 lesions with low-grade dysplasia from those PanIN-2 lesions with moderate-grade dysplasia, each potentially representing a distinct progression step toward invasive carcinoma.
Figures


Comment in
-
Loss of heterozygosity or intragenic mutation, which comes first?Am J Pathol. 2001 May;158(5):1561-3. doi: 10.1016/S0002-9440(10)64109-0. Am J Pathol. 2001. PMID: 11337351 Free PMC article. No abstract available.
References
-
- Cubilla AL, Fitzgerald PJ: Morphological lesions associated with human primary invasive carcinoma nonendocrine pancreas cancer. Cancer Res 1976, 36:2690-2698 - PubMed
-
- Kozuka S, Sassa R, Takai T, Masamoto K, Nagasawa S, Saga S, Hasegawa K, Takeuchi M: Relation of pancreatic duct hyperplasia to carcinoma. Cancer 1979, 43:1418-1428 - PubMed
-
- Klöppel G, Bommer G, Rückert K, Seifert G: Intraductal proliferation in the pancreas and its relationship to human and experimental carcinogenesis. Virchows Arch Pathol Anat 1980, 387:221-233 - PubMed
-
- Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra D, Klöppel G, Longnecker DL, Lüttges J, Offerhaus GJA: Pancreatic intraepithelial neoplasia (PanIN): a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol (in press) - PubMed
-
- Tada M, Ohashi M, Shiratori Y, Okudaira T, Komatsu Y, Kawabe T, Yoshida H, Machinami R: Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology 1996, 110:227-231 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

