Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers
- PMID: 11337385
- PMCID: PMC1891934
- DOI: 10.1016/S0002-9440(10)64143-0
Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers
Abstract
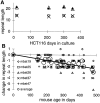
PolyA simple repeat sequence deletions are common in tumors with microsatellite instability (MSI+). Such deletions occur one base at a time in DNA mismatch repair (MMR)-deficient yeast suggesting larger deletions in human MSI+ tumors represent multiple sequential stepwise losses. Sum total deletions in four polyA repeats were variable (between -17 to -45 bp) in 20 sporadic MSI+ colorectal cancers. Progressive but less extensive total deletions (maximum of -12 bp) occurred in similar polyA sequences in MMR-deficient mice (mlh1-/-) up to 478 days old. PolyA repeat lengths were relatively stable but already shortened in the MMR-deficient cell line HCT116. A transgene with 26 A's transfected into HCT116 shortened an average of 3.8 bases pairs after 469 days in culture, less than average deletions of BAT25 (-5.3) or BAT26 (-9.0) in MSI+ cancers. These findings further suggest that extensive polyA deletions common in MSI+ tumors likely reflect multiple stepwise smaller deletions that accumulate more than hundreds of divisions after loss of MMR.
Figures

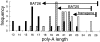
Similar articles
-
Extended microsatellite analysis in microsatellite stable, MSH2 and MLH1 mutation-negative HNPCC patients: genetic reclassification and correlation with clinical features.Digestion. 2004;69(3):166-76. doi: 10.1159/000078223. Epub 2004 Apr 28. Digestion. 2004. PMID: 15118395
-
PolyA deletions in hereditary nonpolyposis colorectal cancer: mutations before a gatekeeper.Am J Pathol. 2002 Apr;160(4):1503-6. doi: 10.1016/S0002-9440(10)62576-X. Am J Pathol. 2002. PMID: 11943734 Free PMC article.
-
Immunohistochemistry and microsatellite instability testing for selecting MLH1, MSH2 and MSH6 mutation carriers in hereditary non-polyposis colorectal cancer.Oncol Rep. 2004 Sep;12(3):621-9. Oncol Rep. 2004. PMID: 15289847
-
[Molecular biology of colorectal cancer--Genetic abnormality: an aspect for an MSI pathway].Nihon Rinsho. 2003 Sep;61 Suppl 7:81-5. Nihon Rinsho. 2003. PMID: 14574860 Review. Japanese. No abstract available.
-
Slip slidin' away: a duodecennial review of targeted genes in mismatch repair deficient colorectal cancer.Crit Rev Oncog. 2007 Dec;13(3):229-57. doi: 10.1615/critrevoncog.v13.i3.20. Crit Rev Oncog. 2007. PMID: 18298386 Review.
Cited by
-
Deep sequencing and pathway-focused analysis revealed multigene oncodriver signatures predicting survival outcomes in advanced colorectal cancer.Oncogenesis. 2018 Jul 22;7(7):55. doi: 10.1038/s41389-018-0066-2. Oncogenesis. 2018. PMID: 30032163 Free PMC article.
-
The clinical significance of PD-L1 in advanced gastric cancer is dependent on ARID1A mutations and ATM expression.Oncoimmunology. 2018 Apr 24;7(8):e1457602. doi: 10.1080/2162402X.2018.1457602. eCollection 2018. Oncoimmunology. 2018. PMID: 30221053 Free PMC article.
-
Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing.J Mol Diagn. 2008 Jul;10(4):301-7. doi: 10.2353/jmoldx.2008.080062. Epub 2008 Jun 13. J Mol Diagn. 2008. PMID: 18556776 Free PMC article.
-
Evolving approach and clinical significance of detecting DNA mismatch repair deficiency in colorectal carcinoma.Semin Diagn Pathol. 2015 Sep;32(5):352-61. doi: 10.1053/j.semdp.2015.02.018. Epub 2015 Feb 4. Semin Diagn Pathol. 2015. PMID: 25716099 Free PMC article. Review.
-
DNA mismatch repair deficiency accelerates endometrial tumorigenesis in Pten heterozygous mice.Am J Pathol. 2002 Apr;160(4):1481-6. doi: 10.1016/S0002-9440(10)62573-4. Am J Pathol. 2002. PMID: 11943731 Free PMC article.
References
-
- Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 - PubMed
-
- Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, Zheng B, Gordon M, Reneker J, Arnheim N, Shibata D, Bradley A, Liskay RM: Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet 1998, 18:276-279 - PubMed
-
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998, 58:5248-5257 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

