Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites
- PMID: 11340174
- PMCID: PMC87046
- DOI: 10.1128/MCB.21.11.3830-3839.2001
Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites
Abstract
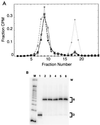
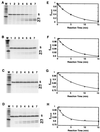
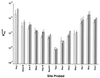
Polypurine tracts are important elements of eukaryotic promoters. They are believed to somehow destabilize chromatin, but the mechanism of their action is not known. We show that incorporating an A(16) element at an end of the nucleosomal DNA and further inward destabilizes histone-DNA interactions by 0.1 +/- 0.03 and 0.35 +/- 0.04 kcal mol(-1), respectively, and is accompanied by 1.5- +/- 0.1-fold and 1.7- +/- 0.1-fold increases in position-averaged equilibrium accessibility of nucleosomal DNA target sites. These effects are comparable in magnitude to effects of A(16) elements that correlate with transcription in vivo, suggesting that our system may capture most of their physiological role. These results point to two distinct but interrelated models for the mechanism of action of polypurine tract promoter elements in vivo. Given a nucleosome positioned over a promoter region, the presence of a polypurine tract in that nucleosome's DNA decreases the stability of the DNA wrapping, increasing the equilibrium accessibility of other DNA target sites buried inside that nucleosome. Alternatively (if nucleosomes are freely mobile), the presence of a polypurine tract provides a free energy bias for the nucleosome to move to alternative locations, thereby changing the equilibrium accessibilities of other nearby DNA target sites.
Figures


References
-
- Anderson J D, Lowary P T, Widom J. Effects of histone acetylation on the dynamic equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2001;307:977–985. - PubMed
-
- Anderson J D, Widom J. Sequence- and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. - PubMed
-
- Cao H, Widlund H R, Simonsson T, Kubista M. TGGA-repeats impair nucleosome formation. J Mol Biol. 1998;281:253–260. - PubMed
-
- Chen W, Tabor S, Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987;50:1047–1055. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
