Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA
- PMID: 11340175
- PMCID: PMC87048
- DOI: 10.1128/MCB.21.11.3840-3852.2001
Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA
Abstract
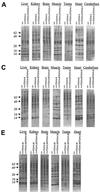
Selenocysteine (Sec) tRNA (tRNA([Ser]Sec)) serves as both the site of Sec biosynthesis and the adapter molecule for donation of this amino acid to protein. The consequences on selenoprotein biosynthesis of overexpressing either the wild type or a mutant tRNA([Ser]Sec) lacking the modified base, isopentenyladenosine, in its anticodon loop were examined by introducing multiple copies of the corresponding tRNA([Ser]Sec) genes into the mouse genome. Overexpression of wild-type tRNA([Ser]Sec) did not affect selenoprotein synthesis. In contrast, the levels of numerous selenoproteins decreased in mice expressing isopentenyladenosine-deficient (i(6)A(-)) tRNA([Ser]Sec) in a protein- and tissue-specific manner. Cytosolic glutathione peroxidase and mitochondrial thioredoxin reductase 3 were the most and least affected selenoproteins, while selenoprotein expression was most and least affected in the liver and testes, respectively. The defect in selenoprotein expression occurred at translation, since selenoprotein mRNA levels were largely unaffected. Analysis of the tRNA([Ser]Sec) population showed that expression of i(6)A(-) tRNA([Ser]Sec) altered the distribution of the two major isoforms, whereby the maturation of tRNA([Ser]Sec) by methylation of the nucleoside in the wobble position was repressed. The data suggest that the levels of i(6)A(-) tRNA([Ser]Sec) and wild-type tRNA([Ser]Sec) are regulated independently and that the amount of wild-type tRNA([Ser]Sec) is determined, at least in part, by a feedback mechanism governed by the level of the tRNA([Ser]Sec) population. This study marks the first example of transgenic mice engineered to contain functional tRNA transgenes and suggests that i(6)A(-) tRNA([Ser]Sec) transgenic mice will be useful in assessing the biological roles of selenoproteins.
Figures




Similar articles
-
Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34.J Biol Chem. 2007 Nov 9;282(45):32591-602. doi: 10.1074/jbc.M707036200. Epub 2007 Sep 11. J Biol Chem. 2007. PMID: 17848557
-
Overproduction of selenocysteine tRNA in Chinese hamster ovary cells following transfection of the mouse tRNA[Ser]Sec gene.RNA. 1998 Nov;4(11):1436-43. doi: 10.1017/s1355838298981043. RNA. 1998. PMID: 9814763 Free PMC article.
-
Inhibition of selenoprotein synthesis by selenocysteine tRNA[Ser]Sec lacking isopentenyladenosine.J Biol Chem. 2000 Sep 8;275(36):28110-9. doi: 10.1074/jbc.M001280200. J Biol Chem. 2000. PMID: 10821829
-
Understanding the role of tRNA modifications in UGA recoding as selenocysteine in eukaryotes.J Mol Biol. 2025 Aug 15;437(16):169017. doi: 10.1016/j.jmb.2025.169017. Epub 2025 Feb 21. J Mol Biol. 2025. PMID: 39988117 Review.
-
Selenocysteine tRNAs as central components of selenoprotein biosynthesis in eukaryotes.Biomed Environ Sci. 1997 Sep;10(2-3):116-24. Biomed Environ Sci. 1997. PMID: 9315302 Review.
Cited by
-
Does a role for selenium in DNA damage repair explain apparent controversies in its use in chemoprevention?Mutagenesis. 2013 Mar;28(2):127-34. doi: 10.1093/mutage/ges064. Epub 2012 Nov 30. Mutagenesis. 2013. PMID: 23204505 Free PMC article. Review.
-
Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice.Antioxid Redox Signal. 2010 Apr 1;12(7):829-38. doi: 10.1089/ars.2009.2895. Antioxid Redox Signal. 2010. PMID: 19769460 Free PMC article.
-
Selenoprotein deficiency accelerates prostate carcinogenesis in a transgenic model.Proc Natl Acad Sci U S A. 2006 May 23;103(21):8179-84. doi: 10.1073/pnas.0508218103. Epub 2006 May 11. Proc Natl Acad Sci U S A. 2006. PMID: 16690748 Free PMC article.
-
Roles for selenium and selenoprotein P in the development, progression, and prevention of intestinal disease.Free Radic Biol Med. 2018 Nov 1;127:26-35. doi: 10.1016/j.freeradbiomed.2018.05.066. Epub 2018 May 17. Free Radic Biol Med. 2018. PMID: 29778465 Free PMC article. Review.
-
Paradoxical Roles of Antioxidant Enzymes: Basic Mechanisms and Health Implications.Physiol Rev. 2016 Jan;96(1):307-64. doi: 10.1152/physrev.00010.2014. Physiol Rev. 2016. PMID: 26681794 Free PMC article. Review.
References
-
- Arner E S, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. - PubMed
-
- Behne D, Hilmet H, Scheid S, Gessner H, Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim Biophys Acta. 1988;996:12–21. - PubMed
-
- Berry M J, Kieffer J D, Harney J W, Larsen P R. Selenocysteine confers the biochemical properties characteristic of the type I iodothyronine deiodinase. J Biol Chem. 1991;266:14155–14158. - PubMed
-
- Bjork G R. Modified nucleosides at positions 34 and 37 of tRNAs and their predicted coding capacities. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: American Society for Microbiology; 1998. pp. 577–581.
-
- Bjork G R, Rasmuson T. Links between tRNA modification and metabolism and modified nucleosides as tumor markers. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: American Society for Microbiology; 1998. pp. 471–492.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
