Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize
- PMID: 11340182
- PMCID: PMC135560
- DOI: 10.1105/tpc.13.5.1063
Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize
Abstract
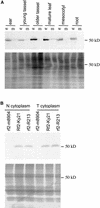
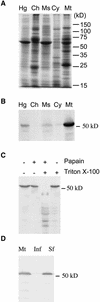
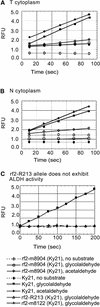
Some plant cytoplasms express novel mitochondrial genes that cause male sterility. Nuclear genes that disrupt the accumulation of the corresponding mitochondrial gene products can restore fertility to such plants. The Texas (T) cytoplasm mitochondrial genome of maize expresses a novel protein, URF13, which is necessary for T cytoplasm-induced male sterility. Working in concert, functional alleles of two nuclear genes, rf1 and rf2, can restore fertility to T cytoplasm plants. Rf1 alleles, but not Rf2 alleles, reduce the accumulation of URF13. Hence, Rf2 differs from typical nuclear restorers in that it does not alter the accumulation of the mitochondrial protein necessary for T cytoplasm-induced male sterility. This study established that the rf2 gene encodes a soluble protein that accumulates in the mitochondrial matrix. Three independent lines of evidence establish that the RF2 protein is an aldehyde dehydrogenase (ALDH). The finding that T cytoplasm plants that are homozygous for the rf2-R213 allele are male sterile but accumulate normal amounts of RF2 protein that lacks normal mitochondrial (mt) ALDH activity provides strong evidence that rf2-encoded mtALDH activity is required to restore male fertility to T cytoplasm maize. Detailed genetic analyses have established that the rf2 gene also is required for anther development in normal cytoplasm maize. Hence, it appears that the rf2 gene was recruited recently to function as a nuclear restorer. ALDHs typically have very broad substrate specificities. Indeed, the RF2 protein is capable of oxidizing at least three aldehydes. Hence, the specific metabolic pathway(s) within which the rf2-encoded mtALDH acts remains to be discovered.
Figures





References
-
- Asker, H., and Davies, D.D. (1985). Mitochondrial aldehyde dehydrogenase from plants. Phytochemistry 24 689–693.
-
- Caballero, E., Baldoma, L., Ros, J., Boronat, A., and Aguilar, J. (1983). Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrogenase as functions of the same protein in Escherichia coli. J. Biol. Chem. 258 7788–7792. - PubMed
-
- Comstock, J.C., and Scheffer, R.P. (1973). Role of host-selective toxin in colonization of corn leaves by Helminthosporium carbonum. Phytopathology 63 24–29.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

