Phosphorylation of murine homeodomain protein Dlx3 by protein kinase C
- PMID: 11343707
- PMCID: PMC1283141
- DOI: 10.1016/s0014-5793(01)02398-5
Phosphorylation of murine homeodomain protein Dlx3 by protein kinase C
Abstract
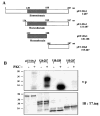
The Dlx3 homeodomain gene is expressed in terminally differentiated murine epidermal cells. As demonstrated for differentiation-specific granular markers, Dlx3 is activated in primary mouse keratinocytes cultured in vitro by increasing the level of the extracellular Ca(2+). This activation is mediated through a protein kinase C-dependent (PKC) pathway. In this study, we investigated whether PKC can modulate the activity of murine Dlx3 protein. Using in vitro kinase assays, we show that PKC enzymes phosphorylate the Dlx3 protein. Using keratinocyte nuclear extracts for the kinase reaction, we determined that Dlx3 protein is phosphorylated, and the phosphorylation is inhibited by the PKC-specific inhibitor GF109203X, suggesting that Dlx3 is phosphorylated by PKC in vivo. Of the PKC isoforms present in the epidermis, we tested alpha, delta, epsilon and zeta. Dlx3 is primarily phosphorylated by PKC alpha. By deletion and mutational analysis, we show that the serine residue S(138), located in the homeodomain of Dlx3 protein, was specifically phosphorylated by PKC. The phosphorylation of purified Dlx3 proteins by PKC partially inhibited formation of complexes between Dlx3 protein and DNA. These results suggest that Dlx3 protein can be directly phosphorylated by PKC and this affects the DNA binding activity of Dlx3.
Figures


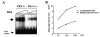
Similar articles
-
Bone morphogenetic protein-2 (BMP-2) transactivates Dlx3 through Smad1 and Smad4: alternative mode for Dlx3 induction in mouse keratinocytes.Nucleic Acids Res. 2002 Jan 15;30(2):515-22. doi: 10.1093/nar/30.2.515. Nucleic Acids Res. 2002. PMID: 11788714 Free PMC article.
-
A novel DLX3-PKC integrated signaling network drives keratinocyte differentiation.Cell Death Differ. 2017 Apr;24(4):717-730. doi: 10.1038/cdd.2017.5. Epub 2017 Feb 10. Cell Death Differ. 2017. PMID: 28186503 Free PMC article.
-
The Dlx3 protein harbors basic residues required for nuclear localization, transcriptional activity and binding to Msx1.J Cell Sci. 2000 Nov;113 ( Pt 22)(Pt 22):4013-23. doi: 10.1242/jcs.113.22.4013. J Cell Sci. 2000. PMID: 11058088 Free PMC article.
-
Cross-talk between epidermal growth factor receptor and protein kinase C during calcium-induced differentiation of keratinocytes.Exp Dermatol. 2000 Jun;9(3):192-9. doi: 10.1034/j.1600-0625.2000.009003192.x. Exp Dermatol. 2000. PMID: 10839717
-
The alpha and eta isoforms of protein kinase C stimulate transcription of human involucrin gene.J Invest Dermatol. 1998 Mar;110(3):218-23. doi: 10.1046/j.1523-1747.1998.00110.x. J Invest Dermatol. 1998. PMID: 9506439 Review.
Cited by
-
Cell-specific phosphorylation of Zfhep transcription factor.Biochem Biophys Res Commun. 2002 Aug 16;296(2):368-73. doi: 10.1016/s0006-291x(02)00880-x. Biochem Biophys Res Commun. 2002. PMID: 12163027 Free PMC article.
-
MAST4 regulates stem cell maintenance with DLX3 for epithelial development and amelogenesis.Exp Mol Med. 2024 Jul;56(7):1606-1619. doi: 10.1038/s12276-024-01264-5. Epub 2024 Jul 1. Exp Mol Med. 2024. PMID: 38945953 Free PMC article.
-
Homeodomain protein Dlx3 induces phosphorylation-dependent p63 degradation.Cell Cycle. 2009 Apr 15;8(8):1185-95. doi: 10.4161/cc.8.8.8202. Epub 2009 Apr 16. Cell Cycle. 2009. PMID: 19282665 Free PMC article.
-
The protein kinase C pathway acts through multiple transcription factors to repress gonadotropin-releasing hormone gene expression in hypothalamic GT1-7 neuronal cells.Mol Endocrinol. 2005 Nov;19(11):2769-79. doi: 10.1210/me.2004-0463. Epub 2005 Jun 30. Mol Endocrinol. 2005. PMID: 15994198 Free PMC article.
-
Bone morphogenetic protein-2 (BMP-2) transactivates Dlx3 through Smad1 and Smad4: alternative mode for Dlx3 induction in mouse keratinocytes.Nucleic Acids Res. 2002 Jan 15;30(2):515-22. doi: 10.1093/nar/30.2.515. Nucleic Acids Res. 2002. PMID: 11788714 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

