Regulation of expression of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339
- PMID: 11344152
- PMCID: PMC99642
- DOI: 10.1128/JB.183.11.3436-3446.2001
Regulation of expression of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339
Abstract
A new open reading frame, encoding a putative integrase-like protein, was detected downstream from the six genes of the vanD glycopeptide resistance cluster in Enterococcus faecium BM4339 (B. Casadewall and P. Courvalin, J. Bacteriol. 181:3644-3648, 1999). In this cluster, genes coding for the VanR(D)-VanS(D) two-component regulatory system were cotranscribed from the P(R(D)) promoter, whereas transcription of the vanY(D), vanH(D), vanD, vanX(D), and intD genes was initiated from the P(Y(D)) promoter located between vanS(D) and vanY(D) (the D subscript indicates that the gene is part of the vanD operon). The VanR(D)-VanS(D) regulatory system is likely to activate transcription of the resistance genes from the promoter P(Y(D)). Glycopeptide-susceptible derivatives of BM4339 were obtained by trans complementation of the frameshift mutation in the ddl gene, restoring functional D-alanine:D-alanine ligase activity in this strain. The glycopeptide-susceptible transformant BM4409, producing only D-alanyl-D-alanine-terminating peptidoglycan precursors, did not express the resistance genes encoding the VanY(D) D,D-carboxypeptidase, the VanH(D) dehydrogenase, the VanD ligase, the VanX(D) D,D-dipeptidase, and also the IntD integrase, although the regulatory region of the vanD cluster was still transcribed. In BM4409, the absence of VanR(D)-VanS(D), apparently dependent, transcription from promoter P(Y(D)) correlated with the lack of D-alanyl-D-lactate-terminating precursors. The vanX(D) gene was transcribed in BM4339, but detectable amounts of VanX(D) D,D-dipeptidase were not synthesized. However, the gene directed synthesis of an active enzyme when cloned on a multicopy plasmid in Escherichia coli, suggesting that the enzyme was unstable in BM4339 or that it had very low activity that was detectable only under conditions of high gene dosage. This activity is not required for glycopeptide resistance in BM4339, since this strain cannot synthesize D-alanyl-D-alanine.
Figures
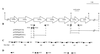
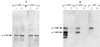
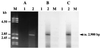




Similar articles
-
VanD-type vancomycin-resistant Enterococcus faecium 10/96A.Antimicrob Agents Chemother. 2003 Jan;47(1):7-18. doi: 10.1128/AAC.47.1.7-18.2003. Antimicrob Agents Chemother. 2003. PMID: 12499162 Free PMC article. Review.
-
VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis.Antimicrob Agents Chemother. 2004 Oct;48(10):3892-904. doi: 10.1128/AAC.48.10.3892-3904.2004. Antimicrob Agents Chemother. 2004. PMID: 15388450 Free PMC article.
-
Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339.J Bacteriol. 1999 Jun;181(12):3644-8. doi: 10.1128/JB.181.12.3644-3648.1999. J Bacteriol. 1999. PMID: 10368136 Free PMC article.
-
New combinations of mutations in VanD-Type vancomycin-resistant Enterococcus faecium, Enterococcus faecalis, and Enterococcus avium strains.Antimicrob Agents Chemother. 2009 May;53(5):1952-63. doi: 10.1128/AAC.01348-08. Epub 2009 Mar 2. Antimicrob Agents Chemother. 2009. PMID: 19258279 Free PMC article.
-
[Regulation mechanism of glycopeptide resistance expression].Nihon Rinsho. 1997 May;55(5):1206-12. Nihon Rinsho. 1997. PMID: 9155176 Review. Japanese.
Cited by
-
VanD-type vancomycin-resistant Enterococcus faecium 10/96A.Antimicrob Agents Chemother. 2003 Jan;47(1):7-18. doi: 10.1128/AAC.47.1.7-18.2003. Antimicrob Agents Chemother. 2003. PMID: 12499162 Free PMC article. Review.
-
Influence of VanD type resistance on activities of glycopeptides in vitro and in experimental endocarditis due to Enterococcus faecium.Antimicrob Agents Chemother. 2003 Nov;47(11):3515-8. doi: 10.1128/AAC.47.11.3515-3518.2003. Antimicrob Agents Chemother. 2003. PMID: 14576110 Free PMC article.
-
First VanD-Type vancomycin-resistant Enterococcus raffinosus isolate.Antimicrob Agents Chemother. 2006 Nov;50(11):3966-7. doi: 10.1128/AAC.00607-06. Epub 2006 Sep 25. Antimicrob Agents Chemother. 2006. PMID: 17000737 Free PMC article. No abstract available.
-
VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis.Antimicrob Agents Chemother. 2004 Oct;48(10):3892-904. doi: 10.1128/AAC.48.10.3892-3904.2004. Antimicrob Agents Chemother. 2004. PMID: 15388450 Free PMC article.
-
Modes and modulations of antibiotic resistance gene expression.Clin Microbiol Rev. 2007 Jan;20(1):79-114. doi: 10.1128/CMR.00015-06. Clin Microbiol Rev. 2007. PMID: 17223624 Free PMC article. Review.
References
-
- Arthur M, Depardieu F, Cabanié L, Reynolds P, Courvalin P. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol Microbiol. 1998;31:819–830. - PubMed
-
- Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. - PubMed
-
- Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

