Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation
- PMID: 11349031
- PMCID: PMC98369
- DOI: 10.1128/IAI.69.6.3685-3691.2001
Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation
Abstract
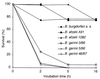
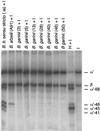
The most characteristic features of the Lyme disease pathogens, the Borrelia burgdorferi sensu lato (s.l.) group, are their ability to invade tissues and to circumvent the immune defenses of the host for extended periods of time, despite elevated levels of borrelia-specific antibodies in serum and other body fluids. Our aim in the present study was to determine whether B. burgdorferi is able to interfere with complement (C) at the level of C3 by accelerating C3b inactivation and thus to inhibit the amplification of the C cascade. Strains belonging to different genospecies (Borrelia garinii, B. burgdorferi sensu stricto, and Borrelia afzelii) were compared for their sensitivities to normal human serum and abilities to promote factor I-mediated C3b degradation. B. burgdorferi sensu stricto and B. afzelii strains were found to be serum resistant. When the spirochetes were incubated with radiolabeled C3b, factor I-mediated degradation of C3b was observed in the presence of C-resistant B. afzelii (n = 3) and B. burgdorferi sensu stricto (n = 1) strains but not in the presence of C-sensitive B. garinii (n = 7) strains or control bacteria (Escherichia coli, Staphylococcus aureus, and Enterococcus faecalis). Immunoblotting and radioligand binding analyses showed that the C-resistant strains had the capacity to acquire the C inhibitors factor H and factor H-like protein 1 (FHL-1) from growth medium and human serum. A novel surface protein with an apparent molecular mass of 35 kDa was found to preferentially bind to the N terminus region of factor H. Thus, the serum-resistant B. burgdorferi s.l. strains can circumvent C attack by binding the C inhibitors factor H and FHL-1 to their surfaces and promoting factor I-mediated C3b degradation.
Figures




References
-
- Anthonissen F M, De Kesel M, Hoet P P, Bigaignon G H. Evidence for the involvement of different genospecies of Borrelia in the clinical outcome of Lyme disease in Belgium. Res Microbiol. 1994;145:327–331. - PubMed
-
- Assous V M, Postic D, Paul G, Nevot P, Baranton G. Western blot analysis of sera from Lyme borreliosis patients according to the genomic species of the Borrelia strains used as antigens. Eur J Clin Microbiol Infect Dis. 1993;12:261–268. - PubMed
-
- Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. - PubMed
-
- Breitner-Ruddock S, Würzner R, Schulze J, Brade V. Heterogeneity in the complement-dependent bacteriolysis within the species of Borrelia burgdorferi. Med Microbiol Immunol. 1997;185:253–260. - PubMed
-
- Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

