26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide
- PMID: 11350924
- PMCID: PMC125470
- DOI: 10.1093/emboj/20.10.2357
26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide
Abstract
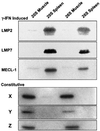
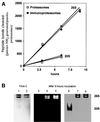
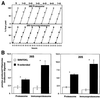
Protein degradation by proteasomes is the source of most antigenic peptides presented on MHC class I molecules. To determine whether proteasomes generate these peptides directly or longer precursors, we developed new methods to measure the efficiency with which 26S and 20S particles, during degradation of a protein, generate the presented epitope or potential precursors. Breakdown of ovalbumin by the 26S and 20S proteasomes yielded the immunodominant peptide SIINFEKL, but produced primarily variants containing 1-7 additional N-terminal residues. Only 6-8% of the times that ovalbumin molecules were digested was a SIINFEKL or an N-extended version produced. Surprisingly, immunoproteasomes which contain the interferon-gamma-induced beta-subunits and are more efficient in antigen presentation, produced no more SIINFEKL than proteasomes. However, the immunoproteasomes released 2-4 times more of certain N-extended versions. These observations show that the changes in cleavage specificity of immunoproteasomes influence not only the C-terminus, but also the N-terminus of potential antigenic peptides, and suggest that most MHC-presented peptides result from N-terminal trimming of larger proteasome products by aminopeptidases (e.g. the interferon-gamma-induced enzyme leucine aminopeptidase).
Figures



References
-
- Akopian T.N., Kisselev,A.F. and Goldberg,A.L. (1997) Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J. Biol. Chem., 272, 1791–1798. - PubMed
-
- Benaroudj N. and Goldberg,A.L. (2000) PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nature Cell Biol., 2, 833–839. - PubMed
-
- Beninga J., Rock,K.L. and Goldberg,A.L. (1998) Interferon-γ can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem., 273, 18734–18742. - PubMed
-
- Ben-Shahar S., Komlosh,A., Nadav,E., Shaked,I., Ziv,T., Admon,A., DeMartino,G.N. and Reiss,Y. (1999) 26 S proteasome-mediated production of an authentic major histocompatibility class I-restricted epitope from an intact protein substrate. J. Biol. Chem., 274, 21963–21972. - PubMed
-
- Cardozo C. and Kohanski,R.A. (1998) Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the ‘immunoproteasome’. J. Biol. Chem., 273, 16764–16770. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

