Functional reconstitution of bacterial Tat translocation in vitro
- PMID: 11350936
- PMCID: PMC125449
- DOI: 10.1093/emboj/20.10.2472
Functional reconstitution of bacterial Tat translocation in vitro
Abstract
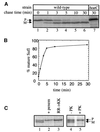
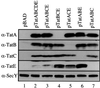
The Tat (twin-arginine translocation) pathway is a Sec-independent mechanism for translocating folded preproteins across or into the inner membrane of Escherichia coli. To study Tat translocation, we sought an in vitro translocation assay using purified inner membrane vesicles and in vitro synthesized substrate protein. While membrane vesicles derived from wild-type cells translocate the Sec-dependent substrate proOmpA, translocation of a Tat-dependent substrate, SufI, was not detected. We established that in vivo overexpression of SufI can saturate the Tat translocase, and that simultaneous overexpression of TatA, B and C relieves this SufI saturation. Using membrane vesicles derived from cells overexpressing TatABC, in vitro translocation of SufI was detected. Like translocation in vivo, translocation of SufI in vitro requires TatABC, an intact membrane potential and the twin-arginine targeting motif within the signal peptide of SUFI: In contrast to Sec translocase, we find that Tat translocase does not require ATP. The development of an in vitro translocation assay is a prerequisite for further biochemical investigations of the mechanism of translocation, substrate recognition and translocase structure.
Figures




References
-
- Bartolome B., Jubete,Y., Martinez,E. and de la Cruz,F. (1991) Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene, 102, 75–78. - PubMed
-
- Berks B.C. (1996) A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol., 22, 393–404. - PubMed
-
- Berks B.C., Sargent,F. and Palmer,T. (2000a) The Tat protein export family. Mol. Microbiol., 35, 260–274. - PubMed
-
- Berks B.C., Sargent,F., De Leeuw,E., Hinsley,A.P., Stanely,N.R., Jack,R.L., Buchanan,G. and Palmer,T. (2000b) A novel transport system involved in the biogenesis of bacterial electron transfer chains. Biochim. Biophys. Acta, 1459, 325–330. - PubMed
-
- Bogsch E.G., Sargent,F., Stanley,N.R., Berks,B.C., Robinson,C. and Palmer,T. (1998) An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem., 273, 18003–18006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

