Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function
- PMID: 11350941
- PMCID: PMC125459
- DOI: 10.1093/emboj/20.10.2518
Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function
Abstract
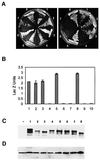
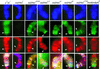
The gypsy insulator is thought to play a role in nuclear organization and the establishment of higher order chromatin domains by bringing together several individual insulator sites to form rosette-like structures in the interphase nucleus. The Su(Hw) and Mod(mdg4) proteins are components of the gypsy insulator required for its effect on enhancer-promoter interactions. Using the yeast two-hybrid system, we show that the Mod(mdg4) protein can form homodimers, which can then interact with Su(Hw). The BTB domain of Mod(mdg4) is involved in homodimerization, whereas the C-terminal region of the protein is involved in interactions with the leucine zipper and adjacent regions of the Su(Hw) protein. Analyses using immunolocalization on polytene chromosomes confirm the involvement of these domains in mediating the interactions between these proteins. Studies using diploid interphase cells further suggest the contribution of these domains to the formation of rosette-like structures in the nucleus. The results provide a biochemical basis for the aggregation of multiple insulator sites and support the role of the gypsy insulator in nuclear organization.
Figures



References
-
- Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. - PubMed
-
- Bell A.C. and Felsenfeld,G. (1999) Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev., 9, 191–198. - PubMed
-
- Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–396. - PubMed
-
- Bell A.C., West,A.G. and Felsenfeld,G. (2001) Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science, 291, 447–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

