The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development
- PMID: 11351070
- PMCID: PMC102281
- DOI: 10.1104/pp.126.1.47
The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development
Abstract
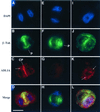
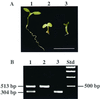
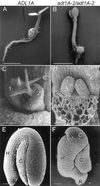
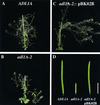
Dynamin and dynamin-like proteins are GTP-binding proteins involved in vesicle trafficking. In soybean, a 68-kD dynamin-like protein called phragmoplastin has been shown to be associated with the cell plate in dividing cells (Gu and Verma, 1996). Five ADL1 genes encoding dynamin-like proteins related to phragmoplastin have been identified in the completed Arabidopsis genome. Here we report that ADL1Ap is associated with punctate subcellular structures and with the cell plate in dividing cells. To assess the function of ADL1Ap we utilized a reverse genetic approach to isolate three separate Arabidopsis mutant lines containing T-DNA insertions in ADL1A. Homozygous adl1A seeds were shriveled and mutant seedlings arrested soon after germination, producing only two leaf primordia and severely stunted roots. Immunoblotting revealed that ADL1Ap expression was not detectable in the mutants. Despite the loss of ADL1Ap, the mutants did not display any defects in cytokinesis, and growth of the mutant seedlings could be rescued in tissue culture by the addition of sucrose. Although these sucrose-rescued plants displayed normal vegetative growth and flowered, they set very few seeds. Thus, ADL1Ap is critical for several stages of plant development, including embryogenesis, seedling development, and reproduction. We discuss the putative role of ADL1Ap in vesicular trafficking, cytokinesis, and other aspects of plant growth.
Figures


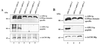


References
-
- Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet. 1996;253:267–277. - PubMed
-
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128.
-
- Baumlein H, Nagy I, Villarroel R, Inzé D, Wobus U. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992;2:233–239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

