Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants
- PMID: 11351099
- PMCID: PMC102310
- DOI: 10.1104/pp.126.1.363
Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants
Abstract
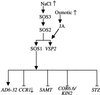
Repetitive rounds of differential subtraction screening, followed by nucleotide sequence determination and northern-blot analysis, identified 84 salt-regulated (160 mM NaCl for 4 h) genes in Arabidopsis wild-type (Col-0 gl1) seedlings. Probes corresponding to these 84 genes and ACP1, RD22BP1, MYB2, STZ, and PAL were included in an analysis of salt responsive gene expression profiles in gl1 and the salt-hypersensitive mutant sos3. Six of 89 genes were expressed differentially in wild-type and sos3 seedlings; steady-state mRNA abundance of five genes (AD06C08/unknown, AD05E05/vegetative storage protein 2 [VSP2], AD05B11/S-adenosyl-L-Met:salicylic acid carboxyl methyltransferase [SAMT], AD03D05/cold regulated 6.6/inducible2 [COR6.6/KIN2], and salt tolerance zinc finger [STZ]) was induced and the abundance of one gene (AD05C10/circadian rhythm-RNA binding1 [CCR1]) was reduced in wild-type plants after salt treatment. The expression of CCR1, SAMT, COR6.6/KIN2, and STZ was higher in sos3 than in wild type, and VSP2 and AD06C08/unknown was lower in the mutant. Salt-induced expression of VSP2 in sos1 was similar to wild type, and AD06C08/unknown, CCR1, SAMT, COR6.6/KIN2, and STZ were similar to sos3. VSP2 is regulated presumably by SOS2/3 independent of SOS1, whereas the expression of the others is SOS1 dependent. AD06C08/unknown and VSP2 are postulated to be effectors of salt tolerance whereas CCR1, SAMT, COR6.6/KIN2, and STZ are determinants that must be negatively regulated during salt adaptation. The pivotal function of the SOS signal pathway to mediate ion homeostasis and salt tolerance implicates AD06C08/unknown, VSP2, SAMT, 6.6/KIN2, STZ, and CCR1 as determinates that are involved in salt adaptation.
Figures


References
-
- Berger S, Bell E, Sadka A, Mullet JE. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol. 1995;27:933–942. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

