Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family
- PMID: 11353082
- PMCID: PMC55449
- DOI: 10.1093/nar/29.10.2127
Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family
Abstract
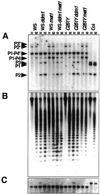
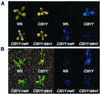
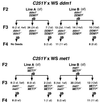
In Arabidopsis a SWI2/SNF2 chromatin remodeling factor-related protein DDM1 and a cytosine methyltransferase MET1 are required for maintenance of genomic cytosine methylation. Mutations in either gene cause global demethylation. In this work we have assessed the effects of these mutations on the PAI tryptophan biosynthetic gene family, which consists of four densely methylated genes arranged as a tail-to-tail inverted repeat plus two unlinked singlet genes. The methylation mutations caused only partial demethylation of the PAI loci: ddm1 had a strong effect on the singlet genes but a weaker effect on the inverted repeat, whereas met1 had a stronger effect on the inverted repeat than on the singlet genes. The double ddm1 met1 mutant also displayed partial demethylation of the PAI genes, with a pattern similar to the ddm1 single mutant. To determine the relationship between partial methylation and expression for the singlet PAI2 gene we constructed a novel reporter strain of Arabidopsis in which PAI2 silencing could be monitored by a blue fluorescent plant phenotype diagnostic of tryptophan pathway defects. This reporter strain revealed that intermediate levels of methylation correlate with intermediate suppression of the fluorescent phenotype.
Figures



References
-
- Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. - PubMed
-
- Finnegan E.J., Genger,R.K., Peacock,W.J. and Dennis,E.S. (1998) DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 223–247. - PubMed
-
- Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E. (1996) De novo cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. - PubMed
-
- Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. - PubMed
-
- Vongs A., Kakutani,T., Martienssen,R.A. and Richards,E.J. (1993) Arabidopsis thaliana DNA methylation mutants. Science, 260, 1926–1928. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

