CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression
- PMID: 11353085
- PMCID: PMC55463
- DOI: 10.1093/nar/29.10.2154
CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression
Abstract
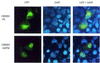
The expression of liver-specific genes is regulated by unequivocally allocated transcription factors via proper responsible elements within their promoters. We identified a novel transcription factor, CREB-H, and found that its expression was restricted in the liver among 16 human tissues tested. A region of CREB-H exhibited significant homology to the basic leucine zipper (b-Zip) domain of members of the CREB/ATF family: mammalian LZIP and Drosophila BBF-2 that binds to box-B, a Drosophila enhancer modulating the fat-body-specific gene expression. CREB-H contained a hydrophobic region representing a putative transmembrane domain, like LZIP. Constructing a variety of CREB-H fusion proteins with the GAL4 DNA-binding domain disclosed that CREB-H functioned as a transcriptional activator and its N-terminal 149 amino acids accounted for the activation ability. Gel mobility sift assays revealed that CREB-H did not bind to the C/EBP, AP-1 and NF-kappaB elements but specifically bound to CRE and the box-B element. Luciferase reporter assays demonstrated that like BBF-2, CREB-H activated transcription via the box-B element and that a deletion of the putative transmembrane domain increased the activation of reporter expression significantly. Furthermore, a fusion protein of GFP and full-length CREB-H was localized in reticular structures surrounding the nucleus, whereas a fusion protein of GFP and a deletion mutant lacking the putative transmembrane domain was mainly in the nucleus. These findings suggest that CREB-H plays an important role in transcriptional regulation of genes specifically expressed in the liver, and that the putative transmembrane domain may be associated with modulation of its function as the transcriptional activator.
Figures






References
-
- Lassar A.B., Buskin,J.N., Lockshon,D., Davis,R.L., Apone,S., Hauschka,S.D. and Weintraub,H. (1989) MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell, 58, 823–831. - PubMed
-
- Donoghue M., Ernst,H., Wentworth,B., Nadal-Ginard,B. and Rosenthal,N. (1988) A muscle-specific enhancer is located at the 3′ end of the myosin light-chain 1/3 gene locus. Genes Dev., 2, 1779–1790. - PubMed
-
- Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science, 240, 1759–1764. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

