Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray
- PMID: 11353609
- PMCID: PMC90529
- DOI: 10.1128/AAC.45.6.1660-1670.2001
Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray
Abstract
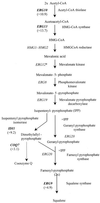
The application of genome-wide expression profiling to determine how drugs achieve their therapeutic effect has provided the pharmaceutical industry with an exciting new tool for drug mode-of-action studies. We used DNA chip technology to study cellular responses to perturbations of ergosterol biosynthesis caused by the broad-spectrum antifungal agent itraconazole. Simultaneous examination of over 6,600 Candida albicans gene transcript levels, representing the entire genome, upon treatment of cells with 10 microM itraconazole revealed that 296 genes were responsive. For 116 genes transcript levels were decreased at least 2.5-fold, while for 180 transcript levels were similarly increased. A global upregulation of ERG genes in response to azole treatment was observed. ERG11 and ERG5 were found to be upregulated approximately 12-fold. In addition, a significant upregulation was observed for ERG6, ERG1, ERG3, ERG4, ERG10, ERG9, ERG26, ERG25, ERG2, IDII, HMGS, NCP1, and FEN2, all of which are genes known to be involved in ergosterol biosynthesis. The effects of itraconazole on a wide variety of known metabolic processes are discussed. As over 140 proteins with unknown function were responsive to itraconazole, our analysis might provide-in combination with phenotypic data-first hints of their potential function. The present report is the first to describe the application of DNA chip technology to study the response of a major human fungal pathogen to drug treatment.
Figures


Comment in
-
Identification of azole-responsive genes by microarray technology: why are we missing the efflux transporter genes?Antimicrob Agents Chemother. 2001 Dec;45(12):3674-6. doi: 10.1128/AAC.45.12.3674-3676.2001. Antimicrob Agents Chemother. 2001. PMID: 11724035 Free PMC article. No abstract available.
Similar articles
-
Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans.Antimicrob Agents Chemother. 2005 Jun;49(6):2226-36. doi: 10.1128/AAC.49.6.2226-2236.2005. Antimicrob Agents Chemother. 2005. PMID: 15917516 Free PMC article.
-
Accumulation of 3-ketosteroids induced by itraconazole in azole-resistant clinical Candida albicans isolates.Antimicrob Agents Chemother. 1999 Nov;43(11):2663-70. doi: 10.1128/AAC.43.11.2663. Antimicrob Agents Chemother. 1999. PMID: 10543744 Free PMC article.
-
Lovastatin synergizes with itraconazole against planktonic cells and biofilms of Candida albicans through the regulation on ergosterol biosynthesis pathway.Appl Microbiol Biotechnol. 2018 Jun;102(12):5255-5264. doi: 10.1007/s00253-018-8959-8. Epub 2018 Apr 25. Appl Microbiol Biotechnol. 2018. PMID: 29691631
-
An update on antifungal targets and mechanisms of resistance in Candida albicans.Med Mycol. 2005 Jun;43(4):285-318. doi: 10.1080/13693780500138971. Med Mycol. 2005. PMID: 16110776 Review.
-
The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn.Virulence. 2016 Aug 17;7(6):649-59. doi: 10.1080/21505594.2016.1188236. Epub 2016 May 24. Virulence. 2016. PMID: 27221657 Free PMC article. Review.
Cited by
-
Sterol C-22 Desaturase ERG5 Mediates the Sensitivity to Antifungal Azoles in Neurospora crassa and Fusarium verticillioides.Front Microbiol. 2013 May 29;4:127. doi: 10.3389/fmicb.2013.00127. eCollection 2013. Front Microbiol. 2013. PMID: 23755044 Free PMC article.
-
Differential gene expression in auristatin PHE-treated Cryptococcus neoformans.Antimicrob Agents Chemother. 2004 Feb;48(2):561-7. doi: 10.1128/AAC.48.2.561-567.2004. Antimicrob Agents Chemother. 2004. PMID: 14742210 Free PMC article.
-
Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria.Antimicrob Agents Chemother. 2004 May;48(5):1600-13. doi: 10.1128/AAC.48.5.1600-1613.2004. Antimicrob Agents Chemother. 2004. PMID: 15105111 Free PMC article.
-
A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates.PLoS Pathog. 2008 Nov;4(11):e1000214. doi: 10.1371/journal.ppat.1000214. Epub 2008 Nov 21. PLoS Pathog. 2008. PMID: 19023412 Free PMC article.
-
The transcription factor Ndt80 does not contribute to Mrr1-, Tac1-, and Upc2-mediated fluconazole resistance in Candida albicans.PLoS One. 2011;6(9):e25623. doi: 10.1371/journal.pone.0025623. Epub 2011 Sep 27. PLoS One. 2011. PMID: 21980509 Free PMC article.
References
-
- Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–7. - PubMed
-
- Daum G, Lees N D, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. - PubMed
-
- Dimster-Denk D, Rine J, Philips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J Lipid Res. 1999;40:850–860. - PubMed
-
- Dixon G, Scanlon D, Cooper S, Broad P. A reporter gene assay for fungal sterol biosynthesis inhibitors. J Steroid Biochem Mol Biol. 1997;62:2–3. , 165–167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

