Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment
- PMID: 11356980
- PMCID: PMC114285
- DOI: 10.1128/JVI.75.12.5703-5710.2001
Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment
Abstract
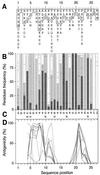
Chronic hepatitis C virus (HCV) infection is a major cause of liver disease. The HCV polyprotein contains a hypervariable region (HVR1) located at the N terminus of the second envelope glycoprotein E2. The strong variability of this 27-amino-acid region is due to its apparent tolerance of amino acid substitutions together with strong selection pressures exerted by anti-HCV immune responses. No specific function has so far been attributed to HVR1. However, its presence at the surface of the viral particle suggests that it might be involved in viral entry. This would imply that HVR1 is not randomly variable. We sequenced 460 HVR1 clones isolated at various times from six HCV-infected patients receiving alpha interferon therapy (which exerts strong pressure towards quasispecies genetic evolution) and analyzed their amino acid sequences together with those of 1,382 nonredundant HVR1 sequences collected from the EMBL database. We found that (i) despite strong amino acid sequence variability related to strong pressures towards change, the chemicophysical properties and conformation of HVR1 were highly conserved, and (ii) HVR1 is a globally basic stretch, with the basic residues located at specific sequence positions. This conservation of positively charged residues indicates that HVR1 is involved in interactions with negatively charged molecules such as lipids, proteins, or glycosaminoglycans (GAGs). As with many other viruses, possible interaction with GAGs probably plays a role in host cell recognition and attachment.
Figures




References
-
- Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. - PubMed
-
- Black S D, Mould D R. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal Biochem. 1991;193:72–82. - PubMed
-
- Blanchet C, Combet C, Geourjon C, Deléage G. MPSA: integrated system for multiple protein sequence analysis with client/server capabilities. Bioinformatics. 2000;16:286–287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

