Cancer chemotherapy--ribonucleases to the rescue
- PMID: 11358688
- PMCID: PMC2913432
- DOI: 10.1016/s1074-5521(01)00030-8
Cancer chemotherapy--ribonucleases to the rescue
Abstract
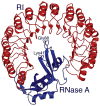
Ribonucleases, once dismissed as uninteresting digestive enzymes, have been shown to have remarkable biological activities. Onconase, from the Northern leopard frog, is currently in clinical trials as a cancer chemotherapeutic. Recent research has revealed some key factors responsible for the cytotoxicity of ribonucleases, and may lead to a new class of drugs.
Figures



Similar articles
-
[Onconase: a ribonuclease with antitumor activity].Postepy Hig Med Dosw (Online). 2010 Feb 19;64:58-66. Postepy Hig Med Dosw (Online). 2010. PMID: 20173221 Review. Polish.
-
Onconase and amphinase, the antitumor ribonucleases from Rana pipiens oocytes.Curr Pharm Biotechnol. 2008 Jun;9(3):215-25. doi: 10.2174/138920108784567245. Curr Pharm Biotechnol. 2008. PMID: 18673287 Free PMC article. Review.
-
Ribonucleases as novel chemotherapeutics : the ranpirnase example.BioDrugs. 2008;22(1):53-8. doi: 10.2165/00063030-200822010-00006. BioDrugs. 2008. PMID: 18215091 Free PMC article. Review.
-
Natural and engineered ribonucleases as potential cancer therapeutics.Biotechnol Lett. 2006 Oct;28(20):1615-22. doi: 10.1007/s10529-006-9145-0. Epub 2006 Aug 11. Biotechnol Lett. 2006. PMID: 16902846 Review.
-
Ribonucleases and immunoRNases as anticancer drugs.Curr Pharm Des. 2009;15(23):2665-75. doi: 10.2174/138161209788923921. Curr Pharm Des. 2009. PMID: 19689337 Review.
Cited by
-
Classical swine fever virus glycoprotein E rns is an endoribonuclease with an unusual base specificity.J Virol. 2004 May;78(10):5507-12. doi: 10.1128/jvi.78.10.5507-5512.2004. J Virol. 2004. PMID: 15113930 Free PMC article.
-
The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants.Mol Cells. 2011 Jan;31(1):25-31. doi: 10.1007/s10059-011-0004-z. Epub 2010 Nov 23. Mol Cells. 2011. PMID: 21110127 Free PMC article.
-
A study of ribonuclease activity in venom of vietnam cobra.J Anim Sci Technol. 2017 Sep 25;59:20. doi: 10.1186/s40781-017-0145-5. eCollection 2017. J Anim Sci Technol. 2017. PMID: 29021904 Free PMC article.
-
Potentiation of ribonuclease cytotoxicity by a poly(amidoamine) dendrimer.Bioorg Med Chem Lett. 2011 May 1;21(9):2756-8. doi: 10.1016/j.bmcl.2010.11.028. Epub 2010 Nov 12. Bioorg Med Chem Lett. 2011. PMID: 21144746 Free PMC article.
-
A Ribonuclease Isolated from Wild Ganoderma Lucidum Suppressed Autophagy and Triggered Apoptosis in Colorectal Cancer Cells.Front Pharmacol. 2016 Jul 25;7:217. doi: 10.3389/fphar.2016.00217. eCollection 2016. Front Pharmacol. 2016. PMID: 27504094 Free PMC article.
References
-
- D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1997.
-
- Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. - PubMed
-
- D’Alessio G. New and cryptic biological messages from RNases. Trends Cell Biol. 1993;3:106–109. - PubMed
-
- Schein CH. From housekeeper to microsurgeon: The diagnostic and therapeutic potential of ribonucleases. Nature Biotechnol. 1997;15:529–536. - PubMed
-
- Riordan JF. Structure and function of angiogenin. In: D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1997. pp. 445–489.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

