NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling
- PMID: 11359904
- PMCID: PMC87059
- DOI: 10.1128/MCB.21.12.3964-3973.2001
NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling
Abstract
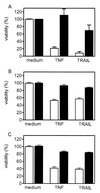
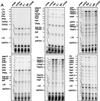
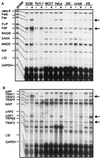
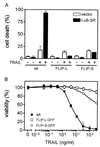
The caspase 8 homologue FLICE-inhibitory protein (cFLIP) is a potent negative regulator of death receptor-induced apoptosis. We found that cFLIP can be upregulated in some cell lines under critical involvement of the NF-kappaB pathway, but NF-kappaB activation was clearly not sufficient for cFLIP induction in all cell lines. Treatment of SV80 cells with the proteasome inhibitor N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal (MG-132) or geldanamycin, a drug interfering with tumor necrosis factor (TNF)-induced NF-kappaB activation, inhibited TNF-induced upregulation of cFLIP. Overexpression of a nondegradable IkappaBalpha mutant (IkappaBalpha-SR) or lack of IkappaB kinase gamma expression completely prevented phorbol myristate acetate-induced upregulation of cFLIP mRNA in Jurkat cells. These data point to an important role for NF-kappaB in the regulation of the cFLIP gene. SV80 cells normally show resistance to TNF-related apoptosis-inducing ligand (TRAIL) and TNF, as apoptosis can be induced only in the presence of low concentrations of cycloheximide (CHX). However, overexpression of IkappaBalpha-SR rendered SV80 cells sensitive to TRAIL-induced apoptosis in the absence of CHX, and cFLIP expression was able to reverse the proapoptotic effect of NF-kappaB inhibition. Western blot analysis further revealed that cFLIP, but not TRAF1, A20, and cIAP2, expression levels rapidly decrease upon CHX treatment. In conclusion, these data suggest a key role for cFLIP in the antiapoptotic response of NF-kappaB activation.
Figures







Similar articles
-
Inhibition of death receptor-mediated gene induction by a cycloheximide-sensitive factor occurs at the level of or upstream of Fas-associated death domain protein (FADD).J Biol Chem. 2000 Aug 11;275(32):24357-66. doi: 10.1074/jbc.M000811200. J Biol Chem. 2000. PMID: 10823821
-
Cellular FLICE/caspase-8-inhibitory protein as a principal regulator of cell death and survival in human hepatocellular carcinoma.Lab Invest. 2003 Jul;83(7):1033-43. doi: 10.1097/01.lab.0000079328.76631.28. Lab Invest. 2003. PMID: 12861043
-
Regulation of tumor necrosis factor-related apoptosis-inducing ligand sensitivity in primary and transformed human keratinocytes.Cancer Res. 2000 Feb 1;60(3):553-9. Cancer Res. 2000. PMID: 10676636
-
Regulation of TRAIL-induced apoptosis by ectopic expression of antiapoptotic factors.Vitam Horm. 2004;67:453-83. doi: 10.1016/S0083-6729(04)67023-3. Vitam Horm. 2004. PMID: 15110190 Review.
-
Mechanisms of resistance to TRAIL-induced apoptosis in cancer.Cancer Gene Ther. 2005 Mar;12(3):228-37. doi: 10.1038/sj.cgt.7700792. Cancer Gene Ther. 2005. PMID: 15550937 Review.
Cited by
-
γ-tocotrienol enhances the chemosensitivity of human oral cancer cells to docetaxel through the downregulation of the expression of NF-κB-regulated anti-apoptotic gene products.Int J Oncol. 2013 Jan;42(1):75-82. doi: 10.3892/ijo.2012.1692. Epub 2012 Nov 8. Int J Oncol. 2013. PMID: 23138939 Free PMC article.
-
USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability.Oncogene. 2017 Jan 26;36(4):458-470. doi: 10.1038/onc.2016.215. Epub 2016 Jun 20. Oncogene. 2017. PMID: 27321185
-
Selenium Deficiency-Induced Oxidative Stress Causes Myocardial Injury in Calves by Activating Inflammation, Apoptosis, and Necroptosis.Antioxidants (Basel). 2023 Jan 19;12(2):229. doi: 10.3390/antiox12020229. Antioxidants (Basel). 2023. PMID: 36829789 Free PMC article.
-
In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8.J Biol Chem. 2011 Apr 15;286(15):13282-91. doi: 10.1074/jbc.M110.216226. Epub 2011 Feb 21. J Biol Chem. 2011. PMID: 21339290 Free PMC article.
-
Keeping Cell Death in Check: Ubiquitylation-Dependent Control of TNFR1 and TLR Signaling.Front Cell Dev Biol. 2019 Jun 28;7:117. doi: 10.3389/fcell.2019.00117. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31316982 Free PMC article.
References
-
- Barkett M, Xue D, Horvitz H R, Gilmore T D. Phosphorylation of IkappaB-alpha inhibits its cleavage by caspase CPP32 in vitro. J Biol Chem. 1997;272:29419–29422. - PubMed
-
- Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. - PubMed
-
- Beg A A, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. - PubMed
-
- Chan F K-M, Lenardo M J. A crucial role for p80 TNF-R2 in amplifying p60 TNF-R1 apoptosis signals in T lymphocytes. Eur J Immunol. 2000;30:652–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
