Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p
- PMID: 11359917
- PMCID: PMC34579
- DOI: 10.1091/mbc.12.5.1215
Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p
Abstract
A family of related proteins in yeast Saccharomyces cerevisiae is known to have in vitro GTPase-activating protein activity on the Rab GTPases. However, their in vivo function remains obscure. One of them, Gyp1p, acts on Sec4p, Ypt1p, Ypt7p, and Ypt51p in vitro. Here, we present data to reveal its in vivo substrate and the role that it plays in the function of the Rab GTPase. Red fluorescent protein-tagged Gyp1p is concentrated on cytoplasmic punctate structures that largely colocalize with a cis-Golgi marker. Subcellular fractionation of a yeast lysate confirmed that Gyp1p is peripherally associated with membranes and that it cofractionates with Golgi markers. This localization suggests that Gyp1p may only act on Rab GTPases on the Golgi. A gyp1Delta strain displays a growth defect on synthetic medium at 37 degrees C. Overexpression of Ypt1p, but not other Rab GTPases, strongly inhibits the growth of gyp1Delta cells. Conversely, a partial loss-of-function allele of YPT1, ypt1-2, can suppress the growth defect of gyp1Delta cells. Furthermore, deletion of GYP1 can partially suppress growth defects associated with mutants in subunits of transport protein particle complex, a complex that catalyzes nucleotide exchange on Ypt1p. These results establish that Gyp1p functions on the Golgi as a negative regulator of Ypt1p.
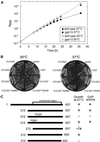
Figures






Similar articles
-
Regulation of membrane traffic by Rab GEF and GAP cascades.Small GTPases. 2016 Oct;7(4):252-256. doi: 10.1080/21541248.2016.1213781. Epub 2016 Jul 18. Small GTPases. 2016. PMID: 27427966 Free PMC article. Review.
-
The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p.Mol Cell Biol. 2004 May;24(9):3815-26. doi: 10.1128/MCB.24.9.3815-3826.2004. Mol Cell Biol. 2004. PMID: 15082776 Free PMC article.
-
Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs.J Biol Chem. 2002 Oct 25;277(43):41023-31. doi: 10.1074/jbc.M205783200. Epub 2002 Aug 19. J Biol Chem. 2002. PMID: 12189143
-
A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14408-13. doi: 10.1073/pnas.0906536106. Epub 2009 Aug 4. Proc Natl Acad Sci U S A. 2009. PMID: 19666511 Free PMC article.
-
[Rab GTPases networks in membrane traffic in Saccharomyces cerevisiae].Yakugaku Zasshi. 2015;135(3):483-92. doi: 10.1248/yakushi.14-00246. Yakugaku Zasshi. 2015. PMID: 25759056 Review. Japanese.
Cited by
-
Are Rab proteins the link between Golgi organization and membrane trafficking?Cell Mol Life Sci. 2012 Dec;69(24):4093-106. doi: 10.1007/s00018-012-1021-6. Epub 2012 May 13. Cell Mol Life Sci. 2012. PMID: 22581368 Free PMC article. Review.
-
Characterization of an M28 metalloprotease family member residing in the yeast vacuole.FEMS Yeast Res. 2013 Aug;13(5):471-84. doi: 10.1111/1567-1364.12050. Epub 2013 Jun 3. FEMS Yeast Res. 2013. PMID: 23679341 Free PMC article.
-
Regulation of membrane traffic by Rab GEF and GAP cascades.Small GTPases. 2016 Oct;7(4):252-256. doi: 10.1080/21541248.2016.1213781. Epub 2016 Jul 18. Small GTPases. 2016. PMID: 27427966 Free PMC article. Review.
-
The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p.Mol Cell Biol. 2004 May;24(9):3815-26. doi: 10.1128/MCB.24.9.3815-3826.2004. Mol Cell Biol. 2004. PMID: 15082776 Free PMC article.
-
Genome-wide screening for genes associated with FK506 sensitivity in fission yeast.PLoS One. 2011;6(8):e23422. doi: 10.1371/journal.pone.0023422. Epub 2011 Aug 5. PLoS One. 2011. PMID: 21850271 Free PMC article.
References
-
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem. 1999;274:33186–33189. - PubMed
-
- Albert S, Gallwitz D. Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol Chem. 2000;381:453–456. - PubMed
-
- Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol. 2000;12:302–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

