NMR and SAXS characterization of the denatured state of the chemotactic protein CheY: implications for protein folding initiation
- PMID: 11369848
- PMCID: PMC2374020
- DOI: 10.1110/ps.52701
NMR and SAXS characterization of the denatured state of the chemotactic protein CheY: implications for protein folding initiation
Abstract
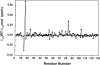
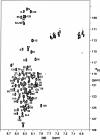
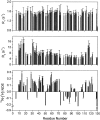
The denatured state of a double mutant of the chemotactic protein CheY (F14N/V83T) has been analyzed in the presence of 5 M urea, using small angle X-ray scattering (SAXS) and heteronuclear magnetic resonance. SAXS studies show that the denatured protein follows a wormlike chain model. Its backbone can be described as a chain composed of rigid elements connected by flexible links. A comparison of the contour length obtained for the chain at 5 M urea with the one expected for a fully expanded chain suggests that approximately 25% of the residues are involved in residual structures. Conformational shifts of the alpha-protons, heteronuclear (15)N-[(1)H] NOEs and (15)N relaxation properties have been used to identify some regions in the protein that deviate from a random coil behavior. According to these NMR data, the protein can be divided into two subdomains, which largely coincide with the two folding subunits identified in a previous kinetic study of the folding of the protein. The first of these subdomains, spanning residues 1-70, is shown here to exhibit a restricted mobility as compared to the rest of the protein. Two regions, one in each subdomain, were identified as deviating from the random coil chemical shifts. Peptides corresponding to these sequences were characterized by NMR and their backbone (1)H chemical shifts were compared to those in the intact protein under identical denaturing conditions. For the region located in the first subdomain, this comparison shows that the observed deviation from random coil parameters is caused by interactions with the rest of the molecule. The restricted flexibility of the first subdomain and the transient collapse detected in that subunit are consistent with the conclusions obtained by applying the protein engineering method to the characterization of the folding reaction transition state.
Figures


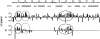

References
-
- Abragam, A. 1961. In The principles of nuclear magnetism. Clarendon Press, Oxford, UK.
-
- Alexandrescu, A.T. and Shortle, D. 1994. Backbone dynamics of a highly disordered 131-residue fragment of staphylococcal nuclease. J. Mol. Biol. 242 527–546. - PubMed
-
- Arcus, V.L., Vuilleumier, S., Freund, S.M.V., Bycroft, M., and Fersht, A.R. 1995. A comparison of the pH, urea and temperature-denatured states of barnase by heteronuclear NMR: Implications for the initiation of protein folding. J. Mol. Biol. 254 305–321. - PubMed
-
- Bax, A. and Davis, D.G. 1985. MLEV-17-based two dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65 355–360.
-
- Blanco, F.J., Serrano, L., and Forman-Kay, J.D. 1998. High populations of non-native structures in the denatured state are compatible with the formation of the native folded state. J. Mol. Biol. 284 1153–1164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

