Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell
- PMID: 11371532
- PMCID: PMC95245
- DOI: 10.1128/JB.183.12.3680-3688.2001
Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell
Abstract
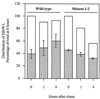
Mycoplasma pneumoniae adsorbs to host respiratory epithelium primarily by its attachment organelle, the proper function of which depends upon mycoplasma adhesin and cytoskeletal proteins. Among the latter are the cytadherence-associated proteins HMW1 and HMW2, whose specific roles in this process are unknown. In the M. pneumoniae cytadherence mutant I-2, loss of HMW2 results in accelerated turnover of HMW1 and other cytadherence-accessory proteins, probably by proteolysis. However, both the mechanism of degradation and the means by which these proteins are rendered susceptible to it are not understood. In this study, we addressed whether HMW1 degradation is a function of its presence among specific subcellular fractions and established that HMW1 is a peripheral membrane protein that is antibody accessible on the outer surfaces of both wild-type and mutant I-2 M. pneumoniae but to a considerably lesser extent in the mutant. Quantitation of HMW1 in Triton X-100-fractionated extracts from cells pulse-labeled with [(35)S]methionine indicated that HMW1 is synthesized in a Triton X-100-soluble form that exists in equilibrium with an insoluble (cytoskeletal) form. Pulse-chase analysis demonstrated that over time, HMW1 becomes stabilized in the cytoskeletal fraction and associated with the cell surface in wild-type M. pneumoniae. The less efficient transition to the cytoskeleton and mycoplasma cell surface in mutant I-2 leads to accelerated degradation of HMW1. These data suggest a role for HMW2 in promoting export of HMW1 to the cell surface, where it is stable and fully functional.
Figures


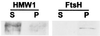

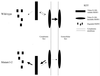

References
-
- Dandekar T, Huynen M, Regula J T, Ueberle B, Zimmermann C U, Andrade M A, Doerks T, Sánchez-Pulido L, Snel B, Suyama M, Yuan Y P, Herrmann R, Bork P. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function, and reading frames. Nucleic Acids Res. 2000;28:3278–3288. - PMC - PubMed
-
- Dirksen L B, Proft T, Hilbert H, Plagens H, Herrmann R, Krause D C. Sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene. 1996;171:19–25. - PubMed
-
- Feldner J, Göbel U, Bredt W. Mycoplasma pneumoniae adhesin is located to the tip structure by monoclonal antibody. Nature (London) 1982;298:765–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

