Bacterial diversity in human subgingival plaque
- PMID: 11371542
- PMCID: PMC95255
- DOI: 10.1128/JB.183.12.3770-3783.2001
Bacterial diversity in human subgingival plaque
Abstract
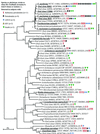
The purpose of this study was to determine the bacterial diversity in the human subgingival plaque by using culture-independent molecular methods as part of an ongoing effort to obtain full 16S rRNA sequences for all cultivable and not-yet-cultivated species of human oral bacteria. Subgingival plaque was analyzed from healthy subjects and subjects with refractory periodontitis, adult periodontitis, human immunodeficiency virus periodontitis, and acute necrotizing ulcerative gingivitis. 16S ribosomal DNA (rDNA) bacterial genes from DNA isolated from subgingival plaque samples were PCR amplified with all-bacterial or selective primers and cloned into Escherichia coli. The sequences of cloned 16S rDNA inserts were used to determine species identity or closest relatives by comparison with sequences of known species. A total of 2,522 clones were analyzed. Nearly complete sequences of approximately 1,500 bases were obtained for putative new species. About 60% of the clones fell into 132 known species, 70 of which were identified from multiple subjects. About 40% of the clones were novel phylotypes. Of the 215 novel phylotypes, 75 were identified from multiple subjects. Known putative periodontal pathogens such as Porphyromonas gingivalis, Bacteroides forsythus, and Treponema denticola were identified from multiple subjects, but typically as a minor component of the plaque as seen in cultivable studies. Several phylotypes fell into two recently described phyla previously associated with extreme natural environments, for which there are no cultivable species. A number of species or phylotypes were found only in subjects with disease, and a few were found only in healthy subjects. The organisms identified only from diseased sites deserve further study as potential pathogens. Based on the sequence data in this study, the predominant subgingival microbial community consisted of 347 species or phylotypes that fall into 9 bacterial phyla. Based on the 347 species seen in our sample of 2,522 clones, we estimate that there are 68 additional unseen species, for a total estimate of 415 species in the subgingival plaque. When organisms found on other oral surfaces such as the cheek, tongue, and teeth are added to this number, the best estimate of the total species diversity in the oral cavity is approximately 500 species, as previously proposed.
Figures






References
-
- Beck J, Garcia R, Heiss G, Vokonas P S, Offenbacher S. Periodontal disease and cardiovascular disease. J Periodontol. 1996;67:1123–1137. - PubMed
-
- Berbari E F, Cockerill III F R, Steckelberg J M. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin Proc. 1997;72:532–542. - PubMed
-
- Boches S K, Mitchell P M, Galvin J L, Loesche W J, Kazor C E, Dewhirst F E, Paster B J. Cultivable and uncultivable bacteria on the healthy tongue dorsum. J Dent Res. 2000;79:396.
-
- Boneh S, Boneh A, Caron R J. Estimating the prediction function and the number of unseen species in sampling with replacement. J Am Statist Assoc. 1998;93:372–379.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

