O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability
- PMID: 11371615
- PMCID: PMC34401
- DOI: 10.1073/pnas.111099998
O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability
Abstract
The posttranslational modification of eukaryotic intracellular proteins by O-linked N-acetylglucosamine (O-GlcNAc) monosaccharides is essential for cell viability, yet its precise functional roles are largely unknown. O-GlcNAc transferase utilizes UDP-GlcNAc, the end product of hexosamine biosynthesis, to catalyze this modification. The availability of UDP-GlcNAc correlates with glycosylation levels of intracellular proteins as well as with transcriptional levels of some genes. Meanwhile, transcription factors and RNA polymerase II can be modified by O-GlcNAc. A linkage between transcription factor O-GlcNAcylation and transcriptional regulation therefore has been postulated. Here, we show that O-GlcNAcylation of a chimeric transcriptional activator containing the second activation domain of Sp1 decreases its transcriptional activity both in an in vitro transcription system and in living cells, which is in concert with our observation that O-GlcNAcylation of Sp1 activation domain blocks its in vitro and in vivo interactions with other Sp1 molecules and TATA-binding protein-associated factor II 110. Furthermore, overexpression of O-GlcNAc transferase specifically inhibits transcriptional activation by native Sp1 in cells. Thus, our studies provide direct evidence that O-GlcNAcylation of transcription factors is involved in transcriptional regulation.
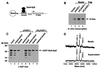
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

