Protein-coding genes are epigenetically regulated in Arabidopsis polyploids
- PMID: 11371624
- PMCID: PMC34425
- DOI: 10.1073/pnas.121064698
Protein-coding genes are epigenetically regulated in Arabidopsis polyploids
Abstract
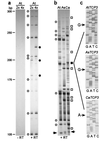
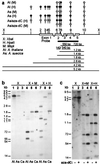
The fate of redundant genes resulting from genome duplication is poorly understood. Previous studies indicated that ribosomal RNA genes from one parental origin are epigenetically silenced during interspecific hybridization or polyploidization. Regulatory mechanisms for protein-coding genes in polyploid genomes are unknown, partly because of difficulty in studying expression patterns of homologous genes. Here we apply amplified fragment length polymorphism (AFLP)-cDNA display to perform a genome-wide screen for orthologous genes silenced in Arabidopsis suecica, an allotetraploid derived from Arabidopsis thaliana and Cardaminopsis arenosa. We identified ten genes that are silenced from either A. thaliana or C. arenosa origin in A. suecica and located in four of the five A. thaliana chromosomes. These genes represent a variety of RNA and predicted proteins including four transcription factors such as TCP3. The silenced genes in the vicinity of TCP3 are hypermethylated and reactivated by blocking DNA methylation, suggesting epigenetic regulation is involved in the expression of orthologous genes in polyploid genomes. Compared with classic genetic mutations, epigenetic regulation may be advantageous for selection and adaptation of polyploid species during evolution and development.
Figures


References
-
- Lewis W H. Polyploidy. New York: Plenum; 1980.
-
- Becak M L, Becak W. Cytogenet Cell Genet. 1998;80:28–33. - PubMed
-
- Stebbins G L. Variation and Evolution in Plants. New York: Columbia Univ. Press; 1950.
-
- Stebbins G L. Chromosomal Evolution in Higher Plants. London: Edward Arnold; 1971.
-
- Masterson J. Science. 1994;264:421–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

