Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis
- PMID: 11375166
- PMCID: PMC92910
- DOI: 10.1128/AEM.67.6.2578-2585.2001
Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis
Abstract
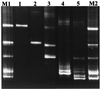
Denaturing gradient gel electrophoresis (DGGE) of DNA fragments generated by PCR with 16S ribosomal DNA-targeted group-specific primers was used to detect lactic acid bacteria (LAB) of the genera Lactobacillus, Pediococcus, Leuconostoc, and Weissella in human feces. Analysis of fecal samples of four subjects revealed individual profiles of DNA fragments originating not only from species that have been described as intestinal inhabitants but also from characteristically food-associated bacteria such as Lactobacillus sakei, Lactobacillus curvatus, Leuconostoc mesenteroides, and Pediococcus pentosaceus. Comparison of PCR-DGGE results with those of bacteriological culture showed that the food-associated species could not be cultured from the fecal samples by plating on Rogosa agar. On the other hand, all of the LAB species cultured from feces were detected in the DGGE profile. We also detected changes in the types of LAB present in human feces during consumption of a milk product containing the probiotic strain Lactobacillus rhamnosus DR20. The analysis of fecal samples from two subjects taken before, during, and after administration of the probiotic revealed that L. rhamnosus was detectable by PCR-DGGE during the test period in the feces of both subjects, whereas it was detectable by culture in only one of the subjects.
Figures


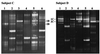

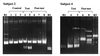
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. - PubMed
-
- Hammes W P, Vogel R F. The genus Lactobacillus. In: Wood B J B, Holzapfel W H, editors. The lactic acid bacteria. 2. The genera of lactic acid bacteria. London, United Kingdom: Blackie Academic and Professional; 1995. pp. 19–54.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

