Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR
- PMID: 11375193
- PMCID: PMC92937
- DOI: 10.1128/AEM.67.6.2766-2774.2001
Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR
Abstract
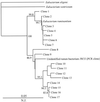
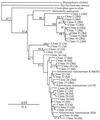
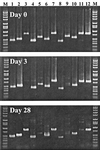
A set of PCR primers was designed and validated for specific detection and quantification of Prevotella ruminicola, Prevotella albensis, Prevotella bryantii, Fibrobacter succinogenes, Selenomonas ruminantium-Mitsuokella multiacida, Streptococcus bovis, Ruminococcus flavefaciens, Ruminobacter amylophilus, Eubacterium ruminantium, Treponema bryantii, Succinivibrio dextrinosolvens, and Anaerovibrio lipolytica. By using these primers and the real-time PCR technique, the corresponding species in the rumens of cows for which the diet was switched from hay to grain were quantitatively monitored. The dynamics of two fibrolytic bacteria, F. succinogenes and R. flavefaciens, were in agreement with those of earlier, culture-based experiments. The quantity of F. succinogenes DNA, predominant in animals on the hay diet, fell 20-fold on the third day of the switch to a grain diet and further declined on day 28, with a 57-fold reduction in DNA. The R. flavefaciens DNA concentration on day 3 declined to approximately 10% of its initial value in animals on the hay diet and remained at this level on day 28. During the transition period (day 3), the quantities of two ruminal prevotella DNAs increased considerably: that of P. ruminicola increased 7-fold and that of P. bryantii increased 263-fold. On day 28, the quantity of P. ruminicola DNA decreased 3-fold, while P. bryantii DNA was still elevated 10-fold in comparison with the level found in animals on the initial hay diet. The DNA specific for another xylanolytic bacterium, E. ruminantium, dropped 14-fold during the diet switch and was maintained at this level on day 28. The concentration of a rumen spirochete, T. bryantii, decreased less profoundly and stabilized with a sevenfold decline by day 28. The variations in A. lipolytica DNA were not statistically significant. After an initial slight increase in S. dextrinosolvens DNA on day 3, this DNA was not detected at the end of the experiment. S. bovis DNA displayed a 67-fold increase during the transition period on day 3. However, on day 28, it actually declined in comparison with the level in animals on the hay ration. The amount of S. ruminantium-M. multiacida DNA also increased eightfold following the diet switch, but stabilized with only a twofold increase on day 28. The real-time PCR technique also uncovered differential amplification of rumen bacterial templates with the set of universal bacterial primers. This observation may explain why some predominant rumen bacteria have not been detected in PCR-generated 16S ribosomal DNA libraries.
Figures


References
-
- Briesacher S L, May T, Grigsby K N, Kerley M S, Anthony R V, Paterson J A. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J Anim Sci. 1992;70:289–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

