Development of an env gp41-based heteroduplex mobility assay for rapid human immunodeficiency virus type 1 subtyping
- PMID: 11376043
- PMCID: PMC88097
- DOI: 10.1128/JCM.39.6.2110-2114.2001
Development of an env gp41-based heteroduplex mobility assay for rapid human immunodeficiency virus type 1 subtyping
Abstract
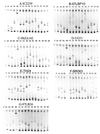
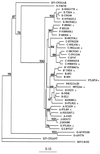
The gp120 region of the human immunodeficiency virus type 1 (HIV-1) envelope (env) gene exhibits a high level of genetic heterogeneity across the group M subtypes. The heteroduplex mobility assay (HMA) has successfully been used to assign subtype classifications, but C2V5 primers often fail to amplify African strains. We developed an env gp41-based HMA for which the target sequence is amplified with highly conserved gp41 primers, known to efficiently amplify nucleic acids from HIV-1 group M, N, and O viruses. By using gp41 from a new panel of reference strains, the subtype assignments made by our modified HMA were concordant with those obtained by sequencing and phylogenetic analysis of 34 field strains from 10 countries representing subtypes A to G. Testing of field strains from Nigeria further demonstrated the utility of this modified assay. Of 28 samples, all could be amplified with gp41 primers but only 17 (60.7%) could be amplified with the standard C2V5 primers. Therefore, gp41-based HMA can be a useful tool for the rapid monitoring of prevalent subtypes in countries with divergent strains of circulating HIV-1.
Figures
References
-
- Bachmann M H, Delwart E L, Shpaer E G, Lingenfelter P, Singal R, Mullins J I. Rapid genetic characterization of HIV type 1 strains from four World Health Organization-sponsored vaccine evaluation sites using a heteroduplex mobility assay. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retrovir. 1994;10:1345–1353. - PubMed
-
- Bobkov A, Cheingsong-Popov R, Garaev M, Rzhaninova A, Kaleebu P, Beddows S, Bachmann M H, Mullins J I, Louwagie J, Janssens W, van der Groen G, Mc Cutchan F, Weber J. Identification of an env G subtype and heterogeneity of HIV-1 strains in the Russian Federation and Belarus. AIDS. 1994;8:1649–1655. - PubMed
-
- Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. - PubMed
-
- Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

