Phylogeny of Pneumocystis carinii from 18 primate species confirms host specificity and suggests coevolution
- PMID: 11376046
- PMCID: PMC88100
- DOI: 10.1128/JCM.39.6.2126-2133.2001
Phylogeny of Pneumocystis carinii from 18 primate species confirms host specificity and suggests coevolution
Abstract
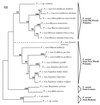
Primates are regularly infected by fungal organisms identified as Pneumocystis carinii. They constitute a valuable population for the confirmation of P. carinii host specificity. In this study, the presence of P. carinii was assessed by direct examination and nested PCR at mitochondrial large subunit (mtLSU) rRNA and dihydropteroate synthetase (DHPS) genes in 98 lung tissue samples from captive or wild nonhuman primates. Fifty-nine air samples corresponding to the environment of different primate species in zoological parks were also examined. Cystic forms of P. carinii were detected in smears from 7 lung tissue samples corresponding to 5 New World primate species. Amplifications at the mtLSU rRNA gene were positive for 29 lung tissue samples representing 18 different primate species or subspecies and 2 air samples corresponding to the environment of two simian colonies. Amplifications at the DHPS gene were positive for 8 lung tissue samples representing 6 different primate species. Direct sequencing of nested PCR products demonstrated that a specific mtLSU rRNA and DHPS sequence could be attributed to each primate species or subspecies. No nonhuman primate harbored the human type of P. carinii (P. carinii f. sp. hominis). Genetic divergence in primate-derived P. carinii organisms varied in terms of the phylogenetic divergence existing among the corresponding host species, suggesting coevolution.
Figures

References
-
- Aliouat E M, Mazars E, Dei-Cas E, Cesbron J Y, Camus D. Intranasal inoculation of mouse, rat or rabbit-derived Pneumocystis in SCID mice. J Protozool Res. 1993;3:94–98.
-
- Aliouat E M, Mazars E, Dei-Cas E, Delcourt P, Billault P, Camus D. Pneumocystis cross infection experiments using SCID mice and nude rats as recipient host showed strong host-species specificity. J Eukaryot Microbiol. 1994;41:S71. - PubMed
-
- Atzori C, Agostoni F, Angeli E, Mainini A, Micheli V, Cargnel A. Pneumocystis carinii host specificity: attempt of cross infection with human derived strains in rats. J Eukaryot Microbiol. 1999;46:S112. - PubMed
-
- Aviles P, Aliouat E M, Martinez A, Dei-Cas E, Herreros E, Dujardin L, Gargallo-Viola D. In vitro pharmacodynamic parameters of sordarin derivatives in comparison with those of marketed compounds against Pneumocystis carinii isolated from rats. Antimicrob Agents Chemother. 2000;44:1284–1290. - PMC - PubMed
-
- Banerji S, Wakefield A E, Alien A G, Maskell D J, Peters S E, Hopkin J M. The cloning and characterization of the arom gene of Pneumocystis carinii. J Gen Microbiol. 1993;139:2901–2914. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

