Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins
- PMID: 11376138
- PMCID: PMC55702
- DOI: 10.1093/nar/29.11.2205
Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins
Abstract
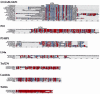
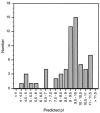
Similarity between the DNA substrates and products of integrase-mediated site-specific recombination reactions results in a single recombinase enzyme being able to catalyze both the integration and excision reactions. The control of directionality in these reactions is achieved through a class of small accessory factors that favor one reaction while interfering with the other. These proteins, which we will refer to collectively as recombination directionality factors (RDFs), play architectural roles in reactions catalyzed by their cognate recombinases and have been identified in conjunction with both tyrosine and serine integrases. Previously identified RDFs are typically small, basic and have diverse amino acid sequences. A subset of RDFs, the cox genes, also function as transcriptional regulators. We present here a compilation of all the known RDF proteins as well as those identified through database mining that we predict to be involved in conferring recombination directionality. Analysis of this group of proteins shows that they can be grouped into distinct sub-groups based on their sequence similarities and that they are likely to have arisen from several independent evolutionary lineages. This compilation will prove useful in recognizing new proteins that confer directionality upon site-specific recombination reactions encoded by plasmids, transposons, phages and prophages.
Figures

References
-
- Nash H.A. (1981) Integration and excision of bacteriophage Lambda: the mechanism of conservation site specific recombination. Annu. Rev. Genet., 15, 143–167. - PubMed
-
- Martin C., Mazodier,P., Mediola,M.V., Gicquel,B., Smokvina,T., Thompson,C.J. and Davies,J. (1991) Site-specific integration of the Streptomyces plasmid pSAM2 in Mycobacterium smegmatis. Mol. Microbiol., 5, 2499–2502. - PubMed
-
- Nash H.A. and Robertson,C.A. (1981) Purification and properties of the Escherichia coli protein factor required for Lambda integrative recombination. J. Biol. Chem., 256, 9246–9253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

