Specific detection of DNA and RNA targets using a novel isothermal nucleic acid amplification assay based on the formation of a three-way junction structure
- PMID: 11376166
- PMCID: PMC55724
- DOI: 10.1093/nar/29.11.e54
Specific detection of DNA and RNA targets using a novel isothermal nucleic acid amplification assay based on the formation of a three-way junction structure
Abstract
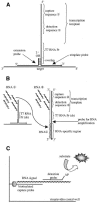
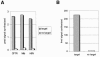
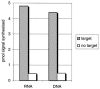
The formation of DNA three-way junction (3WJ) structures has been utilised to develop a novel isothermal nucleic acid amplification assay (SMART) for the detection of specific DNA or RNA targets. The assay consists of two oligonucleotide probes that hybridise to a specific target sequence and, only then, to each other forming a 3WJ structure. One probe (template for the RNA signal) contains a non-functional single-stranded T7 RNA polymerase promoter sequence. This promoter sequence is made double-stranded (hence functional) by DNA polymerase, allowing T7 RNA polymerase to generate a target-dependent RNA signal which is measured by an enzyme-linked oligosorbent assay (ELOSA). The sequence of the RNA signal is always the same, regardless of the original target sequence. The SMART assay was successfully tested in model systems with several single-stranded synthetic targets, both DNA and RNA. The assay could also detect specific target sequences in both genomic DNA and total RNA from Escherichia coli. It was also possible to generate signal from E.coli samples without prior extraction of nucleic acid, showing that for some targets, sample purification may not be required. The assay is simple to perform and easily adaptable to different targets.
Figures



References
-
- Compton J., (1991) Nucleic acid sequence-based amplification. Nature, 350, 91–92. - PubMed
-
- Kievits T., van Gemen,B., van Strijp,D., Schukkink,R., Dircks,M., Adriaanse,H., Malek,L., Sooknanan,R. and Lens,P. (1991) NASBA isothermal enzymatic in vitro nucleic acid amplification optimised for the diagnosis of HIV-1 infection. J. Virol. Methods, 35, 273–286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

