Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea
- PMID: 11381026
- PMCID: PMC311118
- DOI: 10.1101/gr.gr1653r
Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea
Abstract
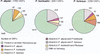
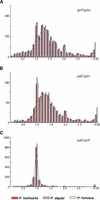
We have compared three complete genomes of closely related hyperthermophilic species of Archaea belonging to the Pyrococcus genus: Pyrococcus abyssi, Pyrococcus horikoshii, and Pyrococcus furiosus. At the genomic level, the comparison reveals a differential conservation among four regions of the Pyrococcus chromosomes correlated with the location of genetic elements mediating DNA reorganization. This discloses the relative contribution of the major mechanisms that promote genomic plasticity in these Archaea, namely rearrangements linked to the replication terminus, insertion sequence-mediated recombinations, and DNA integration within tRNA genes. The combination of these mechanisms leads to a high level of genomic plasticity in these hyperthermophilic Archaea, at least comparable to the plasticity observed between closely related bacteria. At the proteomic level, the comparison of the three Pyrococcus species sheds light on specific selection pressures acting both on their coding capacities and evolutionary rates. Indeed, thanks to two independent methods, the "reciprocal best hits" approach and a new distance ratio analysis, we detect the false orthology relationships within the Pyrococcus lineage. This reveals a high amount of differential gains and losses of genes since the divergence of the three closely related species. The resulting polymorphism is probably linked to an adaptation of these free-living organisms to differential environmental constraints. As a corollary, we delineate the set of orthologous genes shared by the three species, that is, the genes that may characterize the Pyrococcus genus. In this conserved core, the amino acid substitution rate is equal between P. abyssi and P. horikoshii for most of their shared proteins, even for fast-evolving ones. In contrast, strong discrepancies exist among the substitution rates observed in P. furiosus relative to the two other species, which is in disagreement with the molecular clock hypothesis.
Figures

References
-
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, de Jonge BL, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
-
- Andersson JO, Andersson SG. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev. 1999;9:664–671. - PubMed
-
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
