Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification
- PMID: 11381035
- PMCID: PMC311129
- DOI: 10.1101/gr.180501
Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification
Abstract
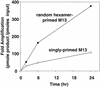
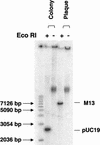
We describe a simple method of using rolling circle amplification to amplify vector DNA such as M13 or plasmid DNA from single colonies or plaques. Using random primers and phi29 DNA polymerase, circular DNA templates can be amplified 10,000-fold in a few hours. This procedure removes the need for lengthy growth periods and traditional DNA isolation methods. Reaction products can be used directly for DNA sequencing after phosphatase treatment to inactivate unincorporated nucleotides. Amplified products can also be used for in vitro cloning, library construction, and other molecular biology applications.
Figures


Similar articles
-
Atomic force microscopy analysis of rolling circle amplification of plasmid DNA.Arch Histol Cytol. 2003 May;66(2):175-81. doi: 10.1679/aohc.66.175. Arch Histol Cytol. 2003. PMID: 12846557
-
Cell-free cloning using multiply-primed rolling circle amplification with modified RNA primers.Biotechniques. 2009 Jul;47(1):609-15. doi: 10.2144/000113155. Biotechniques. 2009. PMID: 19594445
-
Multiple displacement amplification products are compatible with recombination-based cloning.Biotechniques. 2007 Jun;42(6):706, 708. doi: 10.2144/000112496. Biotechniques. 2007. PMID: 17612292 No abstract available.
-
Rolling circle amplification (RCA)-based DNA hydrogel.Nat Protoc. 2021 Dec;16(12):5460-5483. doi: 10.1038/s41596-021-00621-2. Epub 2021 Oct 29. Nat Protoc. 2021. PMID: 34716450 Review.
-
Rolling-circle amplification of viral DNA genomes using phi29 polymerase.Trends Microbiol. 2009 May;17(5):205-11. doi: 10.1016/j.tim.2009.02.004. Epub 2009 Apr 15. Trends Microbiol. 2009. PMID: 19375325 Review.
Cited by
-
Analysis of genomic regions of Trichoderma harzianum IOC-3844 related to biomass degradation.PLoS One. 2015 Apr 2;10(4):e0122122. doi: 10.1371/journal.pone.0122122. eCollection 2015. PLoS One. 2015. PMID: 25836973 Free PMC article.
-
Digital Droplet Multiple Displacement Amplification (ddMDA) for Whole Genome Sequencing of Limited DNA Samples.PLoS One. 2016 May 4;11(5):e0153699. doi: 10.1371/journal.pone.0153699. eCollection 2016. PLoS One. 2016. PMID: 27144304 Free PMC article.
-
Beak and feather disease virus in wild and captive parrots: an analysis of geographic and taxonomic distribution and methodological trends.Arch Virol. 2016 Aug;161(8):2059-74. doi: 10.1007/s00705-016-2871-2. Epub 2016 May 5. Arch Virol. 2016. PMID: 27151279 Free PMC article. Review.
-
Combinatorial library of improved peptide aptamers, CLIPs to inhibit RAGE signal transduction in mammalian cells.PLoS One. 2013 Jun 13;8(6):e65180. doi: 10.1371/journal.pone.0065180. Print 2013. PLoS One. 2013. PMID: 23785412 Free PMC article.
-
Genome-wide detection of single-nucleotide and copy-number variations of a single human cell.Science. 2012 Dec 21;338(6114):1622-6. doi: 10.1126/science.1229164. Science. 2012. PMID: 23258894 Free PMC article.
References
-
- Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. - PubMed
-
- Esteban JA, Salas M, Blanco L. Fidelity of Phi29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. - PubMed
-
- Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols. San Diego: Academic Press; 1990.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
