Nucleolar assembly of the rRNA processing machinery in living cells
- PMID: 11381093
- PMCID: PMC2174343
- DOI: 10.1083/jcb.153.5.1097
Nucleolar assembly of the rRNA processing machinery in living cells
Abstract
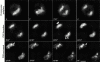
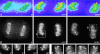
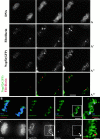
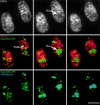
To understand how nuclear machineries are targeted to accurate locations during nuclear assembly, we investigated the pathway of the ribosomal RNA (rRNA) processing machinery towards ribosomal genes (nucleolar organizer regions [NORs]) at exit of mitosis. To follow in living cells two permanently transfected green fluorescence protein-tagged nucleolar proteins, fibrillarin and Nop52, from metaphase to G1, 4-D time-lapse microscopy was used. In early telophase, fibrillarin is concentrated simultaneously in prenucleolar bodies (PNBs) and NORs, whereas PNB-containing Nop52 forms later. These distinct PNBs assemble at the chromosome surface. Analysis of PNB movement does not reveal the migration of PNBs towards the nucleolus, but rather a directional flow between PNBs and between PNBs and the nucleolus, ensuring progressive delivery of proteins into nucleoli. This delivery appeared organized in morphologically distinct structures visible by electron microscopy, suggesting transfer of large complexes. We propose that the temporal order of PNB assembly and disassembly controls nucleolar delivery of these proteins, and that accumulation of processing complexes in the nucleolus is driven by pre-rRNA concentration. Initial nucleolar formation around competent NORs appears to be followed by regroupment of the NORs into a single nucleolus 1 h later to complete the nucleolar assembly. This demonstrates the formation of one functional domain by cooperative interactions between different chromosome territories.
Figures





References
-
- Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. - PubMed
-
- Benavente R. Postmitotic nuclear reorganization events analyzed in living cells. Chromosoma. 1991;100:215–220. - PubMed
-
- Clute P., Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999;1:82–87. - PubMed

