Phi29 family of phages
- PMID: 11381102
- PMCID: PMC99027
- DOI: 10.1128/MMBR.65.2.261-287.2001
Phi29 family of phages
Abstract
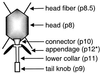
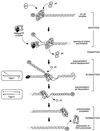
Continuous research spanning more than three decades has made the Bacillus bacteriophage phi29 a paradigm for several molecular mechanisms of general biological processes, such as DNA replication, regulation of transcription, phage morphogenesis, and phage DNA packaging. The genome of bacteriophage phi29 consists of a linear double-stranded DNA (dsDNA), which has a terminal protein (TP) covalently linked to its 5' ends. Initiation of DNA replication, carried out by a protein-primed mechanism, has been studied in detail and is considered to be a model system for the protein-primed DNA replication that is also used by most other linear genomes with a TP linked to their DNA ends, such as other phages, linear plasmids, and adenoviruses. In addition to a continuing progress in unraveling the initiation of DNA replication mechanism and the role of various proteins involved in this process, major advances have been made during the last few years, especially in our understanding of transcription regulation, the head-tail connector protein, and DNA packaging. Recent progress in all these topics is reviewed. In addition to phi29, the genomes of several other Bacillus phages consist of a linear dsDNA with a TP molecule attached to their 5' ends. These phi29-like phages can be divided into three groups. The first group includes, in addition to phi29, phages PZA, phi15, and BS32. The second group comprises B103, Nf, and M2Y, and the third group contains GA-1 as its sole member. Whereas the DNA sequences of the complete genomes of phi29 (group I) and B103 (group II) are known, only parts of the genome of GA-1 (group III) were sequenced. We have determined the complete DNA sequence of the GA-1 genome, which allowed analysis of differences and homologies between the three groups of phi29-like phages, which is included in this review.
Figures


References
-
- Abril A M, Marco S, Carrascosa J L, Salas M, Hermoso J M. Oligomeric structures of the phage φ29 histone-like protein p6. J Mol Biol. 1999;292:581–588. - PubMed
-
- Abril A M, Salas M, Andreu J M, Hermoso J M, Rivas G. Phage φ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentrations found in vivo. Biochemistry. 1997;36:11901–11908. - PubMed
-
- Abril A M, Salas M, Hermoso J M. Identification of residues within two regions involved in self-association of viral histone-like protein p6 from phage φ29. J Biol Chem. 2000;275:26404–26410. - PubMed
-
- Anderson D L, Bodley J W. Role of RNA in bacteriophage φ29 DNA packaging. J Struct Biol. 1990;104:70–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

